Plant Science Research Weekly: August 28, 2020
Review: Extending Plant Defense Theory to Seeds ($)
 Plants have developed multiple mechanisms to deal with the natural enemies they encounter through their life. In consequence, the Plant Defense Theory has arisen to assess how plants allocate resources to this purpose. However, much of the efforts in this matter has revolved around the defense that occurs in saplings and adult plants. In this exciting paper, Dalling et al. synthesize the different defense mechanisms employed by seeds. Since seeds encounter different natural enemies during different stages (i.e., pre-dispersal, post-dispersal, and soil seed banks), the authors discuss the relative importance of chemical and physical defenses in each of these steps. The coordination of defense traits with other seed traits is then discussed, showing that dispersal syndromes and dormancy classes constrain the investment in defenses: while physically dormant seeds would typically invest in physical defenses, the same investment would harm the dispersal capacity of wind-dispersed species. Still, the authors show that it is still unclear if there is a consistent trade-off between seed chemical and physical defenses, or how these defense traits are correlated with the strategies used to defend other organs or the persistence of seeds in the soil. As a result, this review paves the way for future research in the mechanism and evolution of seed defenses. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Annu. Rev. Ecol. Evol. Syst. 10.1146/annurev-ecolsys-012120-115156
Plants have developed multiple mechanisms to deal with the natural enemies they encounter through their life. In consequence, the Plant Defense Theory has arisen to assess how plants allocate resources to this purpose. However, much of the efforts in this matter has revolved around the defense that occurs in saplings and adult plants. In this exciting paper, Dalling et al. synthesize the different defense mechanisms employed by seeds. Since seeds encounter different natural enemies during different stages (i.e., pre-dispersal, post-dispersal, and soil seed banks), the authors discuss the relative importance of chemical and physical defenses in each of these steps. The coordination of defense traits with other seed traits is then discussed, showing that dispersal syndromes and dormancy classes constrain the investment in defenses: while physically dormant seeds would typically invest in physical defenses, the same investment would harm the dispersal capacity of wind-dispersed species. Still, the authors show that it is still unclear if there is a consistent trade-off between seed chemical and physical defenses, or how these defense traits are correlated with the strategies used to defend other organs or the persistence of seeds in the soil. As a result, this review paves the way for future research in the mechanism and evolution of seed defenses. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) Annu. Rev. Ecol. Evol. Syst. 10.1146/annurev-ecolsys-012120-115156
A protein engineering approach for elucidating receptor protein signaling function
 Plant cells express an array of membrane-bound receptor proteins that recognize and respond to extracellular cues. However, their varied structure/specificity and methods of activation pose challenges in assessing their function. Focusing on Leucine-Rich Repeat Receptor Kinases (LRR-RKs), Hohmann et al. used a protein engineering approach to generate chimeras for functional elucidation. Many LRR-RKs require association with a co-receptor SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) to trigger intracellular signaling upon recognition of a ligand. BAK1-INTERACTING RECEPTOR-LIKE KINASES (BIRs) compete with LRR-RKs to bind with the extracellular domain of SERK, independent of any ligands. This competition in effect shuts off the signaling. The authors tried to induce constitutive, ligand-independent expression of several signaling pathways by combining the extracellular and transmembrane domains of a BIR receptor protein (which bind to SERK) and the intracellular kinase domains of different SERK-dependent LRR-RKs, which initiate signaling. These chimeras induced constitutive activation in brassinosteroid signaling, floral abscission, stomatal development, and Casparian strip formation, revealing that this LRR-RK/SERK/BIR interaction is present in a variety of tissues. This “…strategy allows for the identification of gain-of-function phenotypes of orphan LRR-RKs whose ligands are unknown, and enables the elucidation of their receptor activation mechanism.” (Summary by Benjamin Jin) Plant Cell 10.1105/tpc.20.00138
Plant cells express an array of membrane-bound receptor proteins that recognize and respond to extracellular cues. However, their varied structure/specificity and methods of activation pose challenges in assessing their function. Focusing on Leucine-Rich Repeat Receptor Kinases (LRR-RKs), Hohmann et al. used a protein engineering approach to generate chimeras for functional elucidation. Many LRR-RKs require association with a co-receptor SOMATIC EMBRYOGENESIS RECEPTOR-LIKE KINASE (SERK) to trigger intracellular signaling upon recognition of a ligand. BAK1-INTERACTING RECEPTOR-LIKE KINASES (BIRs) compete with LRR-RKs to bind with the extracellular domain of SERK, independent of any ligands. This competition in effect shuts off the signaling. The authors tried to induce constitutive, ligand-independent expression of several signaling pathways by combining the extracellular and transmembrane domains of a BIR receptor protein (which bind to SERK) and the intracellular kinase domains of different SERK-dependent LRR-RKs, which initiate signaling. These chimeras induced constitutive activation in brassinosteroid signaling, floral abscission, stomatal development, and Casparian strip formation, revealing that this LRR-RK/SERK/BIR interaction is present in a variety of tissues. This “…strategy allows for the identification of gain-of-function phenotypes of orphan LRR-RKs whose ligands are unknown, and enables the elucidation of their receptor activation mechanism.” (Summary by Benjamin Jin) Plant Cell 10.1105/tpc.20.00138
The manifold actions of signaling peptides on subcellular dynamics of a receptor specify stomatal cell fate
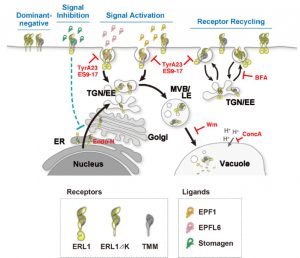 Stomatal development requires cell-to-cell communication and follows one cell spacing, with a minimum of one cell space between two stomata. Key to this communication are the ERECTA and ERECTA LIKE (ERL) Leucine-Rich Repeat domain-Receptor Like kinases (LRR-RLKs) and their ligands, the EPIDERMAL PATTERNING FACTOR (EPF) and EPF-LIKE peptides. Often, a single receptor has different phenotypic outcomes in the same developmental pathway, and how the receptor subcellular dynamics translates to a given phenotype has remained elusive. In this paper, Qi et al. used pharmacological, cell biological, and genetic experiments to investigate the subcellular dynamics of ERL1. Application of the peptides EPF1 and EPFL6, negative regulators of stomatal development, resulted in endocytosis of the ERL1 thorough the trans-Golgi network and multi-vesicular bodies to the vacuole for degradation. ERL1 endocytosis stimulated by EPF1 depends on TOO MANY MOUTHS (TMM), an ERL1 co-receptor, but EPFL6-mediated endocytosis is independent of TMM. However, the interaction of ERL1 with a stimulatory peptide, EPF-LIKE9 (also known as STOMAGEN), results in a high lifetime of the ERL1 on the plasma membrane and accumulation of ERL1 in the endoplasmic reticulum. The authors also show that the cytoplasmic domain of ERL1 is necessary for the ligand-induced subcellular dynamics of ERL1. Thus, this paper shows elegantly how the interaction of different ligands like EPF1, EPFL6 and STOMAGEN to the same receptor have different phenotypic outcomes based on subcellular trafficking. (Summary by Vijaya Batthula @Vijaya_Batthula) eLife 10.7554/eLife.58097
Stomatal development requires cell-to-cell communication and follows one cell spacing, with a minimum of one cell space between two stomata. Key to this communication are the ERECTA and ERECTA LIKE (ERL) Leucine-Rich Repeat domain-Receptor Like kinases (LRR-RLKs) and their ligands, the EPIDERMAL PATTERNING FACTOR (EPF) and EPF-LIKE peptides. Often, a single receptor has different phenotypic outcomes in the same developmental pathway, and how the receptor subcellular dynamics translates to a given phenotype has remained elusive. In this paper, Qi et al. used pharmacological, cell biological, and genetic experiments to investigate the subcellular dynamics of ERL1. Application of the peptides EPF1 and EPFL6, negative regulators of stomatal development, resulted in endocytosis of the ERL1 thorough the trans-Golgi network and multi-vesicular bodies to the vacuole for degradation. ERL1 endocytosis stimulated by EPF1 depends on TOO MANY MOUTHS (TMM), an ERL1 co-receptor, but EPFL6-mediated endocytosis is independent of TMM. However, the interaction of ERL1 with a stimulatory peptide, EPF-LIKE9 (also known as STOMAGEN), results in a high lifetime of the ERL1 on the plasma membrane and accumulation of ERL1 in the endoplasmic reticulum. The authors also show that the cytoplasmic domain of ERL1 is necessary for the ligand-induced subcellular dynamics of ERL1. Thus, this paper shows elegantly how the interaction of different ligands like EPF1, EPFL6 and STOMAGEN to the same receptor have different phenotypic outcomes based on subcellular trafficking. (Summary by Vijaya Batthula @Vijaya_Batthula) eLife 10.7554/eLife.58097
Structural basis of salicylic acid perception by Arabidopsis NPR proteins ($)
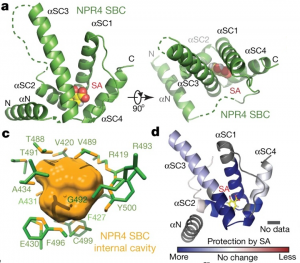 Salicylic acid (SA) is a critical hormone in plant-pathogen responses. The main receptors of this hormonal signal are a small family of NPR proteins (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES). Although NPR1 is a positive regulator of defense signaling, NPR3 and NPR4 serve as negative regulators; they also interact with NPR1 in a SA-mediated manner. Previous efforts to identify the crystal structure of the full-length NPR-SA complex have been unsuccessful. Here, Wang et al. crystalized just the SA-binding core of NPR4 (amino acids 373 to 516) bound to SA. Notably, the structure showed that SA is completely buried within the binding site with no obvious entry point. This finding suggests that there may be a major conformational remodeling of NPR4 upon SA binding. This was confirmed by hydrogen deuterium-exchange mass spectrometry analysis Interestingly, although NPR1 and NPR4 share very similar hormone-binding residues, NPR1 shows much less SA-binding activity when compared to NPR4. Finally, the authors showed that expression of an NPR4 SA hypersensitive variant enhances SA-mediated basal immunity but without affecting its interaction with NPR1 in high levels of SA. With this understanding of SA signaling mechanisms, it could be possible to engineer plants with boosted plant immunity. (Summary by Camila Ribeiro @camicribeiro86) Nature 10.1038/s41586-020-2596-y
Salicylic acid (SA) is a critical hormone in plant-pathogen responses. The main receptors of this hormonal signal are a small family of NPR proteins (NONEXPRESSOR OF PATHOGENESIS-RELATED GENES). Although NPR1 is a positive regulator of defense signaling, NPR3 and NPR4 serve as negative regulators; they also interact with NPR1 in a SA-mediated manner. Previous efforts to identify the crystal structure of the full-length NPR-SA complex have been unsuccessful. Here, Wang et al. crystalized just the SA-binding core of NPR4 (amino acids 373 to 516) bound to SA. Notably, the structure showed that SA is completely buried within the binding site with no obvious entry point. This finding suggests that there may be a major conformational remodeling of NPR4 upon SA binding. This was confirmed by hydrogen deuterium-exchange mass spectrometry analysis Interestingly, although NPR1 and NPR4 share very similar hormone-binding residues, NPR1 shows much less SA-binding activity when compared to NPR4. Finally, the authors showed that expression of an NPR4 SA hypersensitive variant enhances SA-mediated basal immunity but without affecting its interaction with NPR1 in high levels of SA. With this understanding of SA signaling mechanisms, it could be possible to engineer plants with boosted plant immunity. (Summary by Camila Ribeiro @camicribeiro86) Nature 10.1038/s41586-020-2596-y
An interactome of plant autophagy proteins and pathogen effectors ($)
 Autophagy is a regulated process whereby select cytoplasmic components are degraded. It is involved in a variety of biological processes, including defense against pathogens. As such, plant pathogens have evolved mechanisms to target host autophagy machinery to gain virulence. However, we still lack a comprehensive understanding of pathogen molecules that alter plant autophagy pathways. Lal et al. conducted a yeast two-hybrid screen between 25 Arabidopsis autophagy (ATG) proteins and 184 effectors derived from diverse pathogens and found 88 interacting pairs. Among ATG proteins, ATG8 interacted with the highest number of effectors. ATG8 is a central player in autophagy processes such as the promotion of autophagosome biogenesis and the recruitment of specific cargo for degradation. The authors characterized the functions of three effectors of the bacterial pathogen Pseudomonas syringae pv tomato, HrpZ1, HopF1, and AvrPtoB. HrpZ1 and HopF1 both interact with ATG8 but regulate autophagy in opposite directions. Heterologous expression of HrpZ1 or HopF1 in plants conferred susceptibility to a bacterial pathogen. AvrPtoB does not interact with ATG8 but with ATG1, another frequently targeted ATG protein. The authors showed AvrPtoB inhibits autophosphorylation of ATG1, affecting its kinase activity. This study provides a resource that helps researchers to study molecular mechanisms by which pathogens target autophagy processes of plants. (Summary by Tatsuya Nobori @nobolly) Cell Host Microbe 10.1016/j.chom.2020.07.010
Autophagy is a regulated process whereby select cytoplasmic components are degraded. It is involved in a variety of biological processes, including defense against pathogens. As such, plant pathogens have evolved mechanisms to target host autophagy machinery to gain virulence. However, we still lack a comprehensive understanding of pathogen molecules that alter plant autophagy pathways. Lal et al. conducted a yeast two-hybrid screen between 25 Arabidopsis autophagy (ATG) proteins and 184 effectors derived from diverse pathogens and found 88 interacting pairs. Among ATG proteins, ATG8 interacted with the highest number of effectors. ATG8 is a central player in autophagy processes such as the promotion of autophagosome biogenesis and the recruitment of specific cargo for degradation. The authors characterized the functions of three effectors of the bacterial pathogen Pseudomonas syringae pv tomato, HrpZ1, HopF1, and AvrPtoB. HrpZ1 and HopF1 both interact with ATG8 but regulate autophagy in opposite directions. Heterologous expression of HrpZ1 or HopF1 in plants conferred susceptibility to a bacterial pathogen. AvrPtoB does not interact with ATG8 but with ATG1, another frequently targeted ATG protein. The authors showed AvrPtoB inhibits autophosphorylation of ATG1, affecting its kinase activity. This study provides a resource that helps researchers to study molecular mechanisms by which pathogens target autophagy processes of plants. (Summary by Tatsuya Nobori @nobolly) Cell Host Microbe 10.1016/j.chom.2020.07.010
Cell-cell adhesion in plant grafting is facilitated by b-1,4-glucanases ($)
 Plant grafting has been used in crop improvement for centuries and more recently to study systemic and long-distance signaling in the plant vascular system. In order to better understand graft compatibility and its mechanism, Notaguchi et al. used Nicotiana to study interfamily graft combinations. Interfamily grafting between Nicotiana and Chrysanthemum morifolium (Cm, family Asteraceae) was successful and the plants survived through seed setting. By contrast, interfamily grafting between Cm and Glycine max (Gm, family Fabaceae) was not successful and the Gm scions died. A necrotic layer at the graft boundaries in the unsuccessful grafts was observed but was only weakly visible in the successful graft of (Cm/Cm and Nb/Cm). These results suggest that Nicotiana is cable of grafting with phylogenetically distant plant species and demonstrated interfamily cell-cell adhesion. Out of 84 species from 42 families studied, Nicotiana established graft with 73 species from 38 families. Early upregulated genes in the Nb scion were genes encoding cell wall modifying enzymes, suggesting a role of cell wall modification. One of them is NbGH9B3 encoding β-1,4-glucanase. Virus-induced gene silencing of NbGH9B3 caused failure of Nb/At interfamily grafting. CRISPR/Cas9 knock out of NbGH9B3 also led to reduced interfamily grafting in Nb/At, demonstrating that β-1,4-glucanase encoded by NbGH9B3 facilitates graft establishment in Nicotiana interfamily grafting. (Summary by Toluwase Olukayode @toluxylic) Science 10.1126/science.abc3710
Plant grafting has been used in crop improvement for centuries and more recently to study systemic and long-distance signaling in the plant vascular system. In order to better understand graft compatibility and its mechanism, Notaguchi et al. used Nicotiana to study interfamily graft combinations. Interfamily grafting between Nicotiana and Chrysanthemum morifolium (Cm, family Asteraceae) was successful and the plants survived through seed setting. By contrast, interfamily grafting between Cm and Glycine max (Gm, family Fabaceae) was not successful and the Gm scions died. A necrotic layer at the graft boundaries in the unsuccessful grafts was observed but was only weakly visible in the successful graft of (Cm/Cm and Nb/Cm). These results suggest that Nicotiana is cable of grafting with phylogenetically distant plant species and demonstrated interfamily cell-cell adhesion. Out of 84 species from 42 families studied, Nicotiana established graft with 73 species from 38 families. Early upregulated genes in the Nb scion were genes encoding cell wall modifying enzymes, suggesting a role of cell wall modification. One of them is NbGH9B3 encoding β-1,4-glucanase. Virus-induced gene silencing of NbGH9B3 caused failure of Nb/At interfamily grafting. CRISPR/Cas9 knock out of NbGH9B3 also led to reduced interfamily grafting in Nb/At, demonstrating that β-1,4-glucanase encoded by NbGH9B3 facilitates graft establishment in Nicotiana interfamily grafting. (Summary by Toluwase Olukayode @toluxylic) Science 10.1126/science.abc3710
Slow development restores the fertility of photoperiod-sensitive male-sterile plant lines
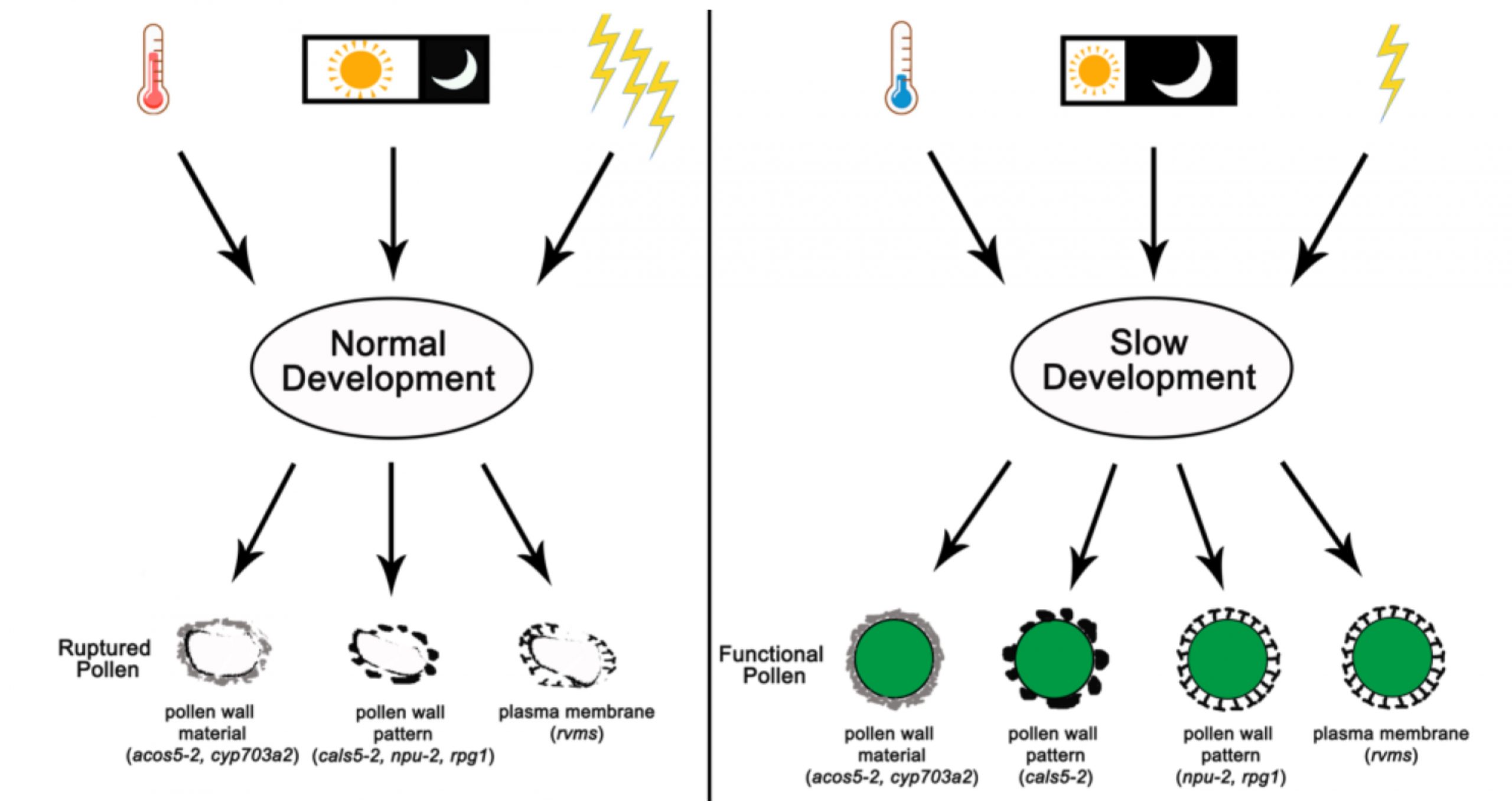 There are many well-known advantages to hybrid seeds. However, one obstacle is the tendency of some plants to self-pollinate. The development of genetically male-sterile lines greatly facilitates hybrid seed production, as the maternal male-sterile plant cannot self-fertilize and depends on donor pollen to set seed. Of course, this raises the question of how these lines are propagated, and one convenient solution is the development of conditional male sterility. Zhang, Xu, Ren et al. investigated a thermo-sensitive conditionally male-sterile Arabidopsis mutant, cals5-2, that has fertility restored at cold temperatures. Interestingly, they found that this mutant is also fertile in short-day or low-light conditions, and that for it and several other temperature-sensitive male-sterile mutants, slowing down the growth rate can overcome the male sterility. Developing pollen cells secrete a complex, multilayer wall, which is abnormal in several of these conditionally sterile mutants. Slow growth restores the fertility of these lines, even with their wall abnormalities. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00951
There are many well-known advantages to hybrid seeds. However, one obstacle is the tendency of some plants to self-pollinate. The development of genetically male-sterile lines greatly facilitates hybrid seed production, as the maternal male-sterile plant cannot self-fertilize and depends on donor pollen to set seed. Of course, this raises the question of how these lines are propagated, and one convenient solution is the development of conditional male sterility. Zhang, Xu, Ren et al. investigated a thermo-sensitive conditionally male-sterile Arabidopsis mutant, cals5-2, that has fertility restored at cold temperatures. Interestingly, they found that this mutant is also fertile in short-day or low-light conditions, and that for it and several other temperature-sensitive male-sterile mutants, slowing down the growth rate can overcome the male sterility. Developing pollen cells secrete a complex, multilayer wall, which is abnormal in several of these conditionally sterile mutants. Slow growth restores the fertility of these lines, even with their wall abnormalities. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00951
DNA damage triggers reprogramming of differentiated cells into stem cells in Physcomitrella ($)
 Wounding and other types of cellular damage, such as DNA damage-induced stem cell death, trigger cellular reprogramming to confer stem cell identity to damage-adjacent cells, thus establishing a new stem cell niche capable of repairing the injury. Studies in angiosperm models have identified the phytohormone auxin and the AP2/ERF transcription factor ERF115 as key players integrating damage responses through cellular reprogramming. A recent study by Gu et al. using the early diverging moss Physcomitrella patens (recently renamed Physcomitrium patens) showed that DNA damage caused by DNA-strand-breaks inducers (i.e. zeocin, bleomycin) causes leaf cell reprogramming to form chloronema apical stem cells, a cell type that’s in an earlier phase of the life cycle than leaf cells. Staining with fluorescent dyes revealed that the observed reprogramming occurs without cell death, while deletion mutant line analyses combined with zeocin treatment demonstrated that DNA-damage induced reprogramming depends on the expression of STEMIN1 (acting redundantly with STEMIN2/STEMIN3), an AP2/ERF transcription factor previously identified as a regulator of reprogramming in response to wounding. Further analyses showed that the expression of the key DNA damage signal transducer ATR (ATM and RAD3-related) is strongly increased after zeocin treatment and is also required for reprogramming, probably through DNA repair or by participating directly in the reprogramming process. This work calls attention to STEMINs as regulators of both wounding and DNA-damage induced reprogramming and opens a new niche for exploration: the relationship between DNA strand breaks and cellular reprogramming in the absence of cell death. (Summary by Jesus Leon @jesussaur) Nature Plants 10.1038/s41477-020-0745-9
Wounding and other types of cellular damage, such as DNA damage-induced stem cell death, trigger cellular reprogramming to confer stem cell identity to damage-adjacent cells, thus establishing a new stem cell niche capable of repairing the injury. Studies in angiosperm models have identified the phytohormone auxin and the AP2/ERF transcription factor ERF115 as key players integrating damage responses through cellular reprogramming. A recent study by Gu et al. using the early diverging moss Physcomitrella patens (recently renamed Physcomitrium patens) showed that DNA damage caused by DNA-strand-breaks inducers (i.e. zeocin, bleomycin) causes leaf cell reprogramming to form chloronema apical stem cells, a cell type that’s in an earlier phase of the life cycle than leaf cells. Staining with fluorescent dyes revealed that the observed reprogramming occurs without cell death, while deletion mutant line analyses combined with zeocin treatment demonstrated that DNA-damage induced reprogramming depends on the expression of STEMIN1 (acting redundantly with STEMIN2/STEMIN3), an AP2/ERF transcription factor previously identified as a regulator of reprogramming in response to wounding. Further analyses showed that the expression of the key DNA damage signal transducer ATR (ATM and RAD3-related) is strongly increased after zeocin treatment and is also required for reprogramming, probably through DNA repair or by participating directly in the reprogramming process. This work calls attention to STEMINs as regulators of both wounding and DNA-damage induced reprogramming and opens a new niche for exploration: the relationship between DNA strand breaks and cellular reprogramming in the absence of cell death. (Summary by Jesus Leon @jesussaur) Nature Plants 10.1038/s41477-020-0745-9
Lipo-chitooligosaccharides as regulatory signals of fungal growth and development
 During symbiosis, the rhizobia bacteria rely on their lipo-chitooligosachharide signals (LCOs) to associate with plants. This signal is perceived by plant receptor like kinase, LysM-containing receptors which activate the common symbiosis signaling pathway (CSSM). Fungal symbiosis with plants has also adopted a similar symbiotic pathway. But, are the LCOs signals limited to symbiotic fungi only? To address this, Rush et al. screened 59 species of fungi (covering most phyla) and found LCOs in exudates from 53 fungal species. Structural characterization of LCOs in exudates was done using mass spectrometry, revealing structural similarity across the fungal kingdom. The authors applied various combinations of synthetic LCOs to a saprophytic human pathogen, Aspergillus fumigatus, which showed not only increase in spore germination but also the transcriptional changes in genes related to cell membrane and perception within 30 minutes. Similar observation of fungal growth was seen in a yeast, Candida glabrata (which didn’t show LCOs exudate). Overall, the authors showed that the LCOs are produced by almost all species of fungi and act as regulatory signals for fungal growth and development. (Summary by Sunita Pathak @psunita980) Nature Comms 10.1038/s41467-020-17615-5
During symbiosis, the rhizobia bacteria rely on their lipo-chitooligosachharide signals (LCOs) to associate with plants. This signal is perceived by plant receptor like kinase, LysM-containing receptors which activate the common symbiosis signaling pathway (CSSM). Fungal symbiosis with plants has also adopted a similar symbiotic pathway. But, are the LCOs signals limited to symbiotic fungi only? To address this, Rush et al. screened 59 species of fungi (covering most phyla) and found LCOs in exudates from 53 fungal species. Structural characterization of LCOs in exudates was done using mass spectrometry, revealing structural similarity across the fungal kingdom. The authors applied various combinations of synthetic LCOs to a saprophytic human pathogen, Aspergillus fumigatus, which showed not only increase in spore germination but also the transcriptional changes in genes related to cell membrane and perception within 30 minutes. Similar observation of fungal growth was seen in a yeast, Candida glabrata (which didn’t show LCOs exudate). Overall, the authors showed that the LCOs are produced by almost all species of fungi and act as regulatory signals for fungal growth and development. (Summary by Sunita Pathak @psunita980) Nature Comms 10.1038/s41467-020-17615-5
Seed predation selects for reproductive variability and synchrony in perennial plants ($)
 Instead of regularly flowering and fruiting each year, some long-lived species exhibit annually variable reproduction events. This phenomenon, known as masting, is considered an adaptation to reduce the losses from seed predation: in years of high reproduction, seeds are so abundant that predators become satiated well before consuming all seeds, while in low production years, predators starve and reduce their density. Still, whether seed predation drives the evolution of masting has seldom been formally tested. Using a database of 12-20 years of observations, Bogdziewicz et al. assessed the relationship between pre-dispersal seed predation and masting behavior (i.e., annual seed production, interannual variability, and among-plant synchrony) in seven plant species. The authors found evidence of selection by pre-dispersal predation in all but two plant species. Despite this, pre-dispersal predation selected different masting characteristics in the other five species, either reproductive synchrony, interannual variability, or both. To explain these differences, the authors discuss that different seed predators with different life histories and characteristics impose different selective pressures on plants. As a result, this research provides an interesting framework for futures studies about the evolution of masting. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.16835
Instead of regularly flowering and fruiting each year, some long-lived species exhibit annually variable reproduction events. This phenomenon, known as masting, is considered an adaptation to reduce the losses from seed predation: in years of high reproduction, seeds are so abundant that predators become satiated well before consuming all seeds, while in low production years, predators starve and reduce their density. Still, whether seed predation drives the evolution of masting has seldom been formally tested. Using a database of 12-20 years of observations, Bogdziewicz et al. assessed the relationship between pre-dispersal seed predation and masting behavior (i.e., annual seed production, interannual variability, and among-plant synchrony) in seven plant species. The authors found evidence of selection by pre-dispersal predation in all but two plant species. Despite this, pre-dispersal predation selected different masting characteristics in the other five species, either reproductive synchrony, interannual variability, or both. To explain these differences, the authors discuss that different seed predators with different life histories and characteristics impose different selective pressures on plants. As a result, this research provides an interesting framework for futures studies about the evolution of masting. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) New Phytol. 10.1111/nph.16835
Synthetic conversion of leaf chloroplasts into carotenoid-rich plastids reveals mechanistic basis of natural chromoplast development ($)
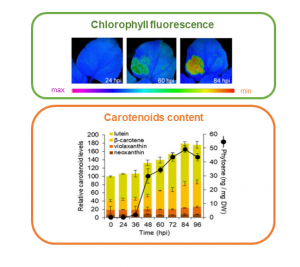 Biofortification aims at increasing the content of health-promoting nutrients in edible parts of the plant. As an example, enhancing the production of carotenoids – natural pigments that provide the yellow to red color – in crops could prevent vitamin A deficiency in humans. In nature, carotenoids accumulate in specialized organelles called chromoplasts, which differentiate from chloroplasts mostly in flowers and fruits, and rarely in leaves. In this article, Llorente and colleagues propose a novel strategy to improve the accumulation of carotenoids in edible green tissues by the means of targeted expression of the bacterial gene CrtB – encoding the enzyme involved in the biosynthesis of phytoene, an intermediate of the carotenoid pathway – in leaf chloroplasts. The CrtB-mediated overaccumulation of phytoene triggers the transformation of leaf chloroplasts into chromoplasts in two steps: reduction of photosynthetic activity, likely mediated by alterations in the internal plastid structures and disassemble of the photosynthetic machinery, and promotion of carotenoid accumulation, likely mediated by upregulation of carotenoid-biosynthetic genes and storage in new structures which prevent their degradation. This transition also requires reprogramming of nuclear gene transcription and primary metabolism. This novel system can be applied to green vegetables to synthetically differentiate chromoplasts from chloroplasts in leaves, and consequently to improve the nutritional values of green organs. (Summary and image preparation by Michela Osnato @michela_osnato) Proc. Natl. Acad. Sci. 10.1073/pnas.2004405117
Biofortification aims at increasing the content of health-promoting nutrients in edible parts of the plant. As an example, enhancing the production of carotenoids – natural pigments that provide the yellow to red color – in crops could prevent vitamin A deficiency in humans. In nature, carotenoids accumulate in specialized organelles called chromoplasts, which differentiate from chloroplasts mostly in flowers and fruits, and rarely in leaves. In this article, Llorente and colleagues propose a novel strategy to improve the accumulation of carotenoids in edible green tissues by the means of targeted expression of the bacterial gene CrtB – encoding the enzyme involved in the biosynthesis of phytoene, an intermediate of the carotenoid pathway – in leaf chloroplasts. The CrtB-mediated overaccumulation of phytoene triggers the transformation of leaf chloroplasts into chromoplasts in two steps: reduction of photosynthetic activity, likely mediated by alterations in the internal plastid structures and disassemble of the photosynthetic machinery, and promotion of carotenoid accumulation, likely mediated by upregulation of carotenoid-biosynthetic genes and storage in new structures which prevent their degradation. This transition also requires reprogramming of nuclear gene transcription and primary metabolism. This novel system can be applied to green vegetables to synthetically differentiate chromoplasts from chloroplasts in leaves, and consequently to improve the nutritional values of green organs. (Summary and image preparation by Michela Osnato @michela_osnato) Proc. Natl. Acad. Sci. 10.1073/pnas.2004405117



