Plant Science Research Weekly: April 24th
Reviews: The physiology of plant responses to drought, and forests and drought ($)
 The increasing global population causes an increasing need for food, but the changing climate means increasing drought occurrences. The April 17 2020 special issue of Science focusses on drought and its effects on food and the environment. The issue includes a fine review by Gupta et al. that provides an overview of how plants are able to sense water limitation and respond to try to avoid dehydration. The authors give an overview of the magnitude of the problem, and how plants respond to mild water stress, for example through changes in root architecture and stomatal responses. They then introduce the signals that effect and coordinate these responses, and discuss possible targets for enhancing them so that crop plants can be developed that are able to produce high yields in spite of water deficit. A second review by Brodribb et al. looks at how trees cope with drought, and the confounding effect elevated temperatures. They discuss water relations in trees, potential for adaptation and acclimation, and modeling approaches to predict drought effects on forests. These fine reviews, written for the broad readership of Science, are excellent choices to share with students. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.aaz7614 and 10.1126/science.aat7631.
The increasing global population causes an increasing need for food, but the changing climate means increasing drought occurrences. The April 17 2020 special issue of Science focusses on drought and its effects on food and the environment. The issue includes a fine review by Gupta et al. that provides an overview of how plants are able to sense water limitation and respond to try to avoid dehydration. The authors give an overview of the magnitude of the problem, and how plants respond to mild water stress, for example through changes in root architecture and stomatal responses. They then introduce the signals that effect and coordinate these responses, and discuss possible targets for enhancing them so that crop plants can be developed that are able to produce high yields in spite of water deficit. A second review by Brodribb et al. looks at how trees cope with drought, and the confounding effect elevated temperatures. They discuss water relations in trees, potential for adaptation and acclimation, and modeling approaches to predict drought effects on forests. These fine reviews, written for the broad readership of Science, are excellent choices to share with students. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.aaz7614 and 10.1126/science.aat7631.
Review: More than a pinch of salt: How plants tolerate salty conditions
 High soil salinity has implications greater than what meets the eye; in addition to the apparent ‘ionic’ stress caused by the toxic sodium (Na+) and chloride (Cl–) ions, it causes osmotic stress (drought-like condition), altered potassium (K+) homeostasis, inhibition of photosynthesis and a plethora of other effects. Plants have evolved complex signaling pathways to deal with these changes and a large number of new pathways have been characterized recently. The last decade provided significant findings including a lipid sensor for salt stress, long-distance calcium (Ca2+) waves, the expansion of the salt overly sensitive (SOS) pathway to include proteins like the flowering time modulator GIGANTEA, etc. Despite many other excellent reviews, van Zelm et al.’s review article is particularly useful, as it spans the length and breadth of salt stress responses from morphological, physiological and metabolic to structural and molecular aspects. It also talks about lesser known aspects and those that need to be ascertained. (Summary by Pavithran Narayanan @pavi_narayanan). Annu. Rev. Plant Biol. 10.1146/annurev-arplant-050718-100005
High soil salinity has implications greater than what meets the eye; in addition to the apparent ‘ionic’ stress caused by the toxic sodium (Na+) and chloride (Cl–) ions, it causes osmotic stress (drought-like condition), altered potassium (K+) homeostasis, inhibition of photosynthesis and a plethora of other effects. Plants have evolved complex signaling pathways to deal with these changes and a large number of new pathways have been characterized recently. The last decade provided significant findings including a lipid sensor for salt stress, long-distance calcium (Ca2+) waves, the expansion of the salt overly sensitive (SOS) pathway to include proteins like the flowering time modulator GIGANTEA, etc. Despite many other excellent reviews, van Zelm et al.’s review article is particularly useful, as it spans the length and breadth of salt stress responses from morphological, physiological and metabolic to structural and molecular aspects. It also talks about lesser known aspects and those that need to be ascertained. (Summary by Pavithran Narayanan @pavi_narayanan). Annu. Rev. Plant Biol. 10.1146/annurev-arplant-050718-100005
Meltome atlas: revealing protein thermal stability across the tree of life
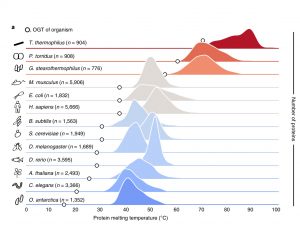 Environmental cues can affect the structure and function of proteins. To get broad empirical information about the effect of temperature on protein stability, Jarzab, Kurzawa, Hopf et al. generated an atlas of proteome thermal stability across 13 model organisms, including bacteria, yeast, worm, fish, fly, plant, mouse and human. After heating cells or lysates, the soluble protein fraction was digested and labeled, followed by peptide identification and quantification using LC-MS/MS, revealing melting features for ~48,000 proteins. With these high-quality thermal proteome profiles, the authors found that in eukaryotes there is an important gap between the optimal growth temperature and the temperature their proteins begin to precipitate (melting temperature). Additionally, they analyzed how factors such as amino acid sequence and composition, protein size and abundance, and protein complex formation affect protein thermal stability. This atlas reports melting points for almost 3000 proteins from Arabidopsis seedlings, with melting temperatures ranging from 30.22 to 68.43 ºC. The curves can be easily visualized and compared in this site: http://meltomeatlas.proteomics.wzw.tum.de:5003. (Summary by Humberto Herrera-Ubaldo @herrera_h Nature Methods 10.1038/s41592-020-0801-4
Environmental cues can affect the structure and function of proteins. To get broad empirical information about the effect of temperature on protein stability, Jarzab, Kurzawa, Hopf et al. generated an atlas of proteome thermal stability across 13 model organisms, including bacteria, yeast, worm, fish, fly, plant, mouse and human. After heating cells or lysates, the soluble protein fraction was digested and labeled, followed by peptide identification and quantification using LC-MS/MS, revealing melting features for ~48,000 proteins. With these high-quality thermal proteome profiles, the authors found that in eukaryotes there is an important gap between the optimal growth temperature and the temperature their proteins begin to precipitate (melting temperature). Additionally, they analyzed how factors such as amino acid sequence and composition, protein size and abundance, and protein complex formation affect protein thermal stability. This atlas reports melting points for almost 3000 proteins from Arabidopsis seedlings, with melting temperatures ranging from 30.22 to 68.43 ºC. The curves can be easily visualized and compared in this site: http://meltomeatlas.proteomics.wzw.tum.de:5003. (Summary by Humberto Herrera-Ubaldo @herrera_h Nature Methods 10.1038/s41592-020-0801-4
AthCNV: A map of DNA copy number variations in the Arabidopsis thaliana genome
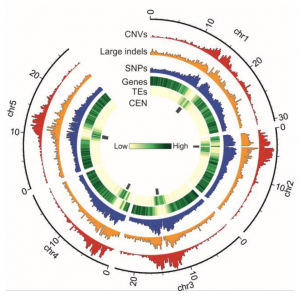 Intraspecific phenotypic variability can be attributed to differences at the genome level such as copy number variations (CNVs). CNVs are large DNA fragments which differ in number between individuals and likely have a crucial evolutionary role. Due to the 1001 Genomes Project, over 1,000 Arabidopsis accessions have been sequenced, with to-date analyses largely focusing on single nucleotide polymorphisms (SNPs) and other small variations. However, there is limited understanding regarding the extent of CNV contribution to genotypic (and phenotypic) diversity. Here, Zmienko et al. generated AthCNV, a map of CNVs in Arabidopsis, which has substantially added to the list of known structural variants. The authors found that CNVs constitute one-third of the genome and are unevenly distributed throughout, with transposable elements overrepresented in these regions. 18.3% of genes fully overlap with CNVs and this group is enriched with genes functioning in defense and stress. To investigate the Arabidopsis population structure, principal component analyses were conducted using CNV markers. Interestingly, the CNV-approach better determined the global distribution and migration patterns of accessions than SNPs. Furthermore, gene copy number losses were more frequent than gains, and 3.9-26.9% of CNV-affected genes were implicated by gene copy number changes in individual accessions. Finally, to link CNVs to phenotypic variation, CNV markers were utilized to perform a genome-wide association study and a strong association was found between Pseudomonas resistance and two CNV-affected R genes. AthCNV has revealed a multitude of previously undetected genomic variants, and this map will facilitate future research to illuminate the impact of CNVs on phenotypic variability in Arabidopsis. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00640
Intraspecific phenotypic variability can be attributed to differences at the genome level such as copy number variations (CNVs). CNVs are large DNA fragments which differ in number between individuals and likely have a crucial evolutionary role. Due to the 1001 Genomes Project, over 1,000 Arabidopsis accessions have been sequenced, with to-date analyses largely focusing on single nucleotide polymorphisms (SNPs) and other small variations. However, there is limited understanding regarding the extent of CNV contribution to genotypic (and phenotypic) diversity. Here, Zmienko et al. generated AthCNV, a map of CNVs in Arabidopsis, which has substantially added to the list of known structural variants. The authors found that CNVs constitute one-third of the genome and are unevenly distributed throughout, with transposable elements overrepresented in these regions. 18.3% of genes fully overlap with CNVs and this group is enriched with genes functioning in defense and stress. To investigate the Arabidopsis population structure, principal component analyses were conducted using CNV markers. Interestingly, the CNV-approach better determined the global distribution and migration patterns of accessions than SNPs. Furthermore, gene copy number losses were more frequent than gains, and 3.9-26.9% of CNV-affected genes were implicated by gene copy number changes in individual accessions. Finally, to link CNVs to phenotypic variation, CNV markers were utilized to perform a genome-wide association study and a strong association was found between Pseudomonas resistance and two CNV-affected R genes. AthCNV has revealed a multitude of previously undetected genomic variants, and this map will facilitate future research to illuminate the impact of CNVs on phenotypic variability in Arabidopsis. (Summary by Caroline Dowling @CarolineD0wling) Plant Cell 10.1105/tpc.19.00640
Evolution of tetraploid meiosis
 Genome duplications are common in plants and thought to be an important contributor to evolutionary innovations, but the increase in ploidy that results from a genome duplication also presents challenges for reproduction. Because there are four sets of homologous chromosomes in the derived tetraploid (instead of two in the diploid ancestor), establishing one-to-one pairing between homologs in meiosis can be a problem. Failure to form such “bivalents” leads to segregation errors and reduced fertility. This would prevent newly formed tetraploids (neopolyploids) from becoming established. However, the plant world is full of polyploid species, meaning that evolution in many cases is able to overcome this obstacle. One way to understand how this could happen is to compare the meiotic division machinery between diploids and tetraploids. Arabidopsis arenosa is an ideal system for this type of investigation because there are both diploid (ancestral) and tetraploid (derived) subpopulations available for comparison. Two meiosis genes, ASY1 and ASY3 are under selection in the tetraploid, suggesting that they may play important roles. These two genes both code for proteins that localize to the so-called chromosome axis, which is central to meiotic pairing. In this paper, Morgan et al. introduce the ASY1 and ASY3 alleles from the diploid ancestor into the tetraploid and notice how this changes several aspects of chromosome behavior during meiotic pairing. In particular, they observe an increased frequency of harmful multivalent pairings. However, the overall effect size was modest. The authors predict that small changes in many genes (including ASY1/3) are needed to cope with the challenges of tetraploid meiosis. (Summary by Frej Tulin @FrejTulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919459117
Genome duplications are common in plants and thought to be an important contributor to evolutionary innovations, but the increase in ploidy that results from a genome duplication also presents challenges for reproduction. Because there are four sets of homologous chromosomes in the derived tetraploid (instead of two in the diploid ancestor), establishing one-to-one pairing between homologs in meiosis can be a problem. Failure to form such “bivalents” leads to segregation errors and reduced fertility. This would prevent newly formed tetraploids (neopolyploids) from becoming established. However, the plant world is full of polyploid species, meaning that evolution in many cases is able to overcome this obstacle. One way to understand how this could happen is to compare the meiotic division machinery between diploids and tetraploids. Arabidopsis arenosa is an ideal system for this type of investigation because there are both diploid (ancestral) and tetraploid (derived) subpopulations available for comparison. Two meiosis genes, ASY1 and ASY3 are under selection in the tetraploid, suggesting that they may play important roles. These two genes both code for proteins that localize to the so-called chromosome axis, which is central to meiotic pairing. In this paper, Morgan et al. introduce the ASY1 and ASY3 alleles from the diploid ancestor into the tetraploid and notice how this changes several aspects of chromosome behavior during meiotic pairing. In particular, they observe an increased frequency of harmful multivalent pairings. However, the overall effect size was modest. The authors predict that small changes in many genes (including ASY1/3) are needed to cope with the challenges of tetraploid meiosis. (Summary by Frej Tulin @FrejTulin) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1919459117
A regulatory module for survival on low potassium
 Potassium ions (K+) play a variety of important roles in plant physiology and are maintained at a high concentration in the cytosol relative to the available K+ in the environment. Potassium may accumulate to an even higher concentration in the vacuole, where it helps to regulate turgor pressure. During periods of potassium scarcity, the plant can increase the cytoplasmic K+ concentration by mobilizing these vacuolar stores. In this paper, Tang et al. describe a pathway that contributes to potassium homeostasis by increasing efflux from the vacuole during low-K+ stress. They show that a specific set of calcium-binding sensor proteins (CBL2 and CBL3) are important for successful germination in low-K+ media, presumably due to increased reliance on internal stores. A similar phenotype resulted from deletion of CBL-interacting protein kinases, especially CIPK9. Current recordings from isolated vacuoles demonstrated that CBLs and CIPKs are indeed important for potassium flux. But how does the CBL-CIPK module change the conductance of the tonoplast towards potassium? One candidate third component is the tonoplast-localized channel TPK1. By co-expressing all three proteins in protoplast vacuoles, they show that the CBL3-CIPK9-TPK1 complex is sufficient to boost potassium efflux, and that this effect is entirely dependent on the kinase activity of CIPK9. Altogether, this study adds new molecular detail to how plants regulate internal potassium levels in response to a changing environment. (Summary by Frej Tulin @FrejTulin) Nature Plants 10.1038/s41477-020-0621-7
Potassium ions (K+) play a variety of important roles in plant physiology and are maintained at a high concentration in the cytosol relative to the available K+ in the environment. Potassium may accumulate to an even higher concentration in the vacuole, where it helps to regulate turgor pressure. During periods of potassium scarcity, the plant can increase the cytoplasmic K+ concentration by mobilizing these vacuolar stores. In this paper, Tang et al. describe a pathway that contributes to potassium homeostasis by increasing efflux from the vacuole during low-K+ stress. They show that a specific set of calcium-binding sensor proteins (CBL2 and CBL3) are important for successful germination in low-K+ media, presumably due to increased reliance on internal stores. A similar phenotype resulted from deletion of CBL-interacting protein kinases, especially CIPK9. Current recordings from isolated vacuoles demonstrated that CBLs and CIPKs are indeed important for potassium flux. But how does the CBL-CIPK module change the conductance of the tonoplast towards potassium? One candidate third component is the tonoplast-localized channel TPK1. By co-expressing all three proteins in protoplast vacuoles, they show that the CBL3-CIPK9-TPK1 complex is sufficient to boost potassium efflux, and that this effect is entirely dependent on the kinase activity of CIPK9. Altogether, this study adds new molecular detail to how plants regulate internal potassium levels in response to a changing environment. (Summary by Frej Tulin @FrejTulin) Nature Plants 10.1038/s41477-020-0621-7
Maize aquaporin PIP2;5 alters water relations and plant growth
 Aquaporins, also known as water channels, are conserved across all kingdoms. In plants, the plasma membrane intrinsic protein (PIP) subfamily controls membrane water permeability. Maize aquaporin PIP2;5 is highly expressed in roots. By reverse genetic approaches, Ding et al. showed that overexpression (OE) and knock-out (KO) of PIP2;5 affect water relations and plant growth upon mild stress. PIP2;5 OE plants show higher hydraulic conductivity of the root cortex cells, with no change in whole root hydraulic conductivity. The quantitative MECHA hydraulic model suggests that the plasma membrane permeability of the cells is not radially uniform, and PIP2;5 could saturate in the endodermis and exodermis layers. Thus, increasing its expression level in the cortex cells has no measurable effect on the conductivity of the whole-root system. By contrast, under mild water stress the time course of leaf elongation rate is faster in the OE lines, where PIP2;5 levels are elevated in leaves relative to wildtype. In the KO lines, conductivity was decreased in the root, but unaffected in the leaves. These results indicate a possible beneficial role for PIP2;5 during stress. (Summary by Min May Wong @wongminmay ) Plant Physiol. 10.1104/pp.19.01183
Aquaporins, also known as water channels, are conserved across all kingdoms. In plants, the plasma membrane intrinsic protein (PIP) subfamily controls membrane water permeability. Maize aquaporin PIP2;5 is highly expressed in roots. By reverse genetic approaches, Ding et al. showed that overexpression (OE) and knock-out (KO) of PIP2;5 affect water relations and plant growth upon mild stress. PIP2;5 OE plants show higher hydraulic conductivity of the root cortex cells, with no change in whole root hydraulic conductivity. The quantitative MECHA hydraulic model suggests that the plasma membrane permeability of the cells is not radially uniform, and PIP2;5 could saturate in the endodermis and exodermis layers. Thus, increasing its expression level in the cortex cells has no measurable effect on the conductivity of the whole-root system. By contrast, under mild water stress the time course of leaf elongation rate is faster in the OE lines, where PIP2;5 levels are elevated in leaves relative to wildtype. In the KO lines, conductivity was decreased in the root, but unaffected in the leaves. These results indicate a possible beneficial role for PIP2;5 during stress. (Summary by Min May Wong @wongminmay ) Plant Physiol. 10.1104/pp.19.01183
Structural evolution drives diversification of the large LRR-RLK gene family
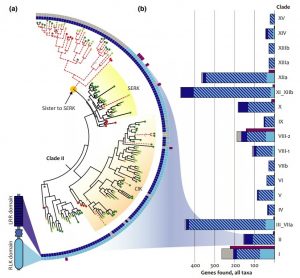 Leucine-rich repeat receptor-like kinases (LRR-RLKs) act as signaling receptors, are the largest plant-specific protein kinase family, and are involved in myriad developmental activities and defense systems. Due to the large number of proteins in this group, their diversification and consequent redundancy have made it difficult to functionally characterize single mutants. To circumvent this issue, Man et al. took a large phylogenetics approach and characterized LRR-RLKs from Arabidopsis thaliana, Amborella trichopoda, Brachypodium distachyon, Oryza sativa, Solanum lycopersicum, Populus trichocarpa, Selaginella moellendorffii, Physcomitrella patens, and Zea mays. This approach revealed new LRR-RLK genes previously uncharacterized, and corrected the classification of this large family of proteins. Most importantly, this work demonstrated the promising way for gene discovery and classification of larger and complex families. (Summary by Arif Ashraf @aribidopsis) New Phytol. 10.1111/nph.16455
Leucine-rich repeat receptor-like kinases (LRR-RLKs) act as signaling receptors, are the largest plant-specific protein kinase family, and are involved in myriad developmental activities and defense systems. Due to the large number of proteins in this group, their diversification and consequent redundancy have made it difficult to functionally characterize single mutants. To circumvent this issue, Man et al. took a large phylogenetics approach and characterized LRR-RLKs from Arabidopsis thaliana, Amborella trichopoda, Brachypodium distachyon, Oryza sativa, Solanum lycopersicum, Populus trichocarpa, Selaginella moellendorffii, Physcomitrella patens, and Zea mays. This approach revealed new LRR-RLK genes previously uncharacterized, and corrected the classification of this large family of proteins. Most importantly, this work demonstrated the promising way for gene discovery and classification of larger and complex families. (Summary by Arif Ashraf @aribidopsis) New Phytol. 10.1111/nph.16455
Mystery of adventitious root formation in grapevine
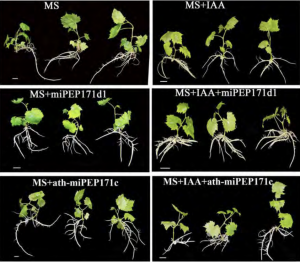 Grapevines (Vitis vinifera L.) are clonally propagated by stem cuttings, which depends on the formation of adventitious (stem-borne) roots. In this paper Chen at al., showed the function of microRNA encoded peptides (miPEPs) in adventitious root formation of cultured grape plantlets. MicroRNA biogenesis involves the transcription of microRNA genes (MIR) by RNA pol II to primary microRNA (pri-miRNA) which is further processed to precursor (pre-miRNA) and mature microRNA (miRNA). In grapevine, miR171 is encoded by 10 genes (vvi-MIR171a to vvi-MIR171j) and targets SCARECROW-LIKE15 and 27 (SCL15, 27) genes, which encode a GRAS domain protein involved in root development. Among these, vvi-MIR171d expression increases, and expression levels of its targets (SCL15, 27) decrease, just before adventitious root formation, indicating the potential role of vvi-MIR171d in root development. The authors identified three small open reading frames (sORF) encoding a potential miPEP in the 500bp upstream of pre-miR171d. Previously it has been shown that in some cases the overexpression of an miPEP can increase the transcription of its own MIR genes. Here, the authors observed increased expression of vvi-MIR171d when one of the sORFs, vvi-miPEP171d1, was overexpressed. Exogenous application of synthetic vvi-miPEP171d1 to the grape tissue culture plantlets increased the adventitious root formation, suggesting a possible use of this peptide in grapevine propagation. However, addition of Arabidopsis thaliana miPEP171c to cultured grape plantlets or vvi-miPEP171d1 to Arabidopsis did not show any effect on root development suggesting a species-specific function. Overall, this study shows the potential use of miPEP in the root development of tissue cultured perennial fruit crops. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant. Physiol. 10.1104/pp.20.00197
Grapevines (Vitis vinifera L.) are clonally propagated by stem cuttings, which depends on the formation of adventitious (stem-borne) roots. In this paper Chen at al., showed the function of microRNA encoded peptides (miPEPs) in adventitious root formation of cultured grape plantlets. MicroRNA biogenesis involves the transcription of microRNA genes (MIR) by RNA pol II to primary microRNA (pri-miRNA) which is further processed to precursor (pre-miRNA) and mature microRNA (miRNA). In grapevine, miR171 is encoded by 10 genes (vvi-MIR171a to vvi-MIR171j) and targets SCARECROW-LIKE15 and 27 (SCL15, 27) genes, which encode a GRAS domain protein involved in root development. Among these, vvi-MIR171d expression increases, and expression levels of its targets (SCL15, 27) decrease, just before adventitious root formation, indicating the potential role of vvi-MIR171d in root development. The authors identified three small open reading frames (sORF) encoding a potential miPEP in the 500bp upstream of pre-miR171d. Previously it has been shown that in some cases the overexpression of an miPEP can increase the transcription of its own MIR genes. Here, the authors observed increased expression of vvi-MIR171d when one of the sORFs, vvi-miPEP171d1, was overexpressed. Exogenous application of synthetic vvi-miPEP171d1 to the grape tissue culture plantlets increased the adventitious root formation, suggesting a possible use of this peptide in grapevine propagation. However, addition of Arabidopsis thaliana miPEP171c to cultured grape plantlets or vvi-miPEP171d1 to Arabidopsis did not show any effect on root development suggesting a species-specific function. Overall, this study shows the potential use of miPEP in the root development of tissue cultured perennial fruit crops. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant. Physiol. 10.1104/pp.20.00197
Innovation, conservation, and repurposing of gene function in plant root cell type development
 Roots have many specialized cells arranged in concentric circles that are functionally homologous among various plant species but with varying cell-type-specific developmental programs. To further understand these developmental programs, Kajala et al. performed TRAPseq (Translating Ribosome Affinity Purification of mRNA coupled with sequencing) to determine differentially expressed translatomes in 12 different cell-types of tomato root. Additionally, the regulators involved in xylem cell differentiation were mapped between tomato and Arabidopsis to determine whether the cell-type regulatory networks are conserved between two different plant species. The author found some conserved transcription factor functions (like HD-ZIPIII), some partially conserved (VND) and some innovative (bZIP11) or repurposed (KNAT1). Root tissue-specific translatome was performed between rice, Arabidopsis, and tomato, which showed extensive meristematic zone similarity between these species with more variation for endodermis, vascular, and meristematic cortex tissue. The authors note that, like in animals, in plants there is a greater conservation amongst gene expression in early developmental stages as compared to differentiated tissues. (Summary by Sunita Pathak @psunita980) bioRxiv 10.1101/2020.04.09.017285
Roots have many specialized cells arranged in concentric circles that are functionally homologous among various plant species but with varying cell-type-specific developmental programs. To further understand these developmental programs, Kajala et al. performed TRAPseq (Translating Ribosome Affinity Purification of mRNA coupled with sequencing) to determine differentially expressed translatomes in 12 different cell-types of tomato root. Additionally, the regulators involved in xylem cell differentiation were mapped between tomato and Arabidopsis to determine whether the cell-type regulatory networks are conserved between two different plant species. The author found some conserved transcription factor functions (like HD-ZIPIII), some partially conserved (VND) and some innovative (bZIP11) or repurposed (KNAT1). Root tissue-specific translatome was performed between rice, Arabidopsis, and tomato, which showed extensive meristematic zone similarity between these species with more variation for endodermis, vascular, and meristematic cortex tissue. The authors note that, like in animals, in plants there is a greater conservation amongst gene expression in early developmental stages as compared to differentiated tissues. (Summary by Sunita Pathak @psunita980) bioRxiv 10.1101/2020.04.09.017285
Trichomes: a means for foliar water uptake (FWU)
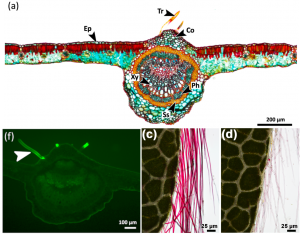 Foliar water uptake (FWU); water uptake through the leaf surface, is observed in several species and helps in drought conditions through water uptake from fog, snow, dew and rain. In this paper, Schreel et al. showed trichomes as a major source of FWU in beech (Fagus sylvatica L.) plants. Trichomes are large epidermal hairy projections on the leaf surface. Using water soluble fluorescent tracers, the authors observed a strong fluorescence in trichomes, a weak signal in epidermal and collenchyma cells, and an absence of signal in substomatal cavities, indicating trichomes as a major means of surface absorption. This is further supported by the evidence that trichomes lack the cuticle and their cell walls are rich in pectin molecules, amenable for water absorption. Beech leaves subjected to 3.5 hours dehydration showed the empty trichomes, of which 75% were filled with water after 4 hours of rehydration. Thus, this work shows trichome-absorption and to a lesser extent cuticular-absorption as means for FWU in beech plants. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant J. 10.1111/tpj.14770
Foliar water uptake (FWU); water uptake through the leaf surface, is observed in several species and helps in drought conditions through water uptake from fog, snow, dew and rain. In this paper, Schreel et al. showed trichomes as a major source of FWU in beech (Fagus sylvatica L.) plants. Trichomes are large epidermal hairy projections on the leaf surface. Using water soluble fluorescent tracers, the authors observed a strong fluorescence in trichomes, a weak signal in epidermal and collenchyma cells, and an absence of signal in substomatal cavities, indicating trichomes as a major means of surface absorption. This is further supported by the evidence that trichomes lack the cuticle and their cell walls are rich in pectin molecules, amenable for water absorption. Beech leaves subjected to 3.5 hours dehydration showed the empty trichomes, of which 75% were filled with water after 4 hours of rehydration. Thus, this work shows trichome-absorption and to a lesser extent cuticular-absorption as means for FWU in beech plants. (Summary by Vijaya Batthula @Vijaya_Batthula) Plant J. 10.1111/tpj.14770
How plants keep their microbiota healthy
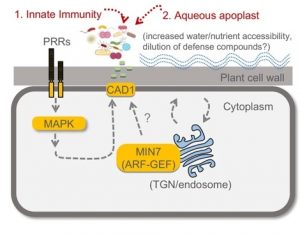 The large apoplastic intercellular space of plant leaves creates nutrient-rich niches for microbial colonization. To date, whether and how plants control the composition of leaf microbiota is poorly understood. Chen et al. reported that the Arabidopsis quadruple mutant (min7fls2efrcerk1 or mfec) defective in pattern-triggered immunity and a vesicle trafficking pathway (MIN7 encodes an ARF-GEF protein) showed tissue damage phenotypes when grown in soils. The mutant plants showed marked changes in the leaf endophytic bacterial community: 1) an overall increased bacterial population size, 2) decreased community diversity, and 3) a shift from a Firmicutes-rich community to a Proteobacteria-rich community. The authors demonstrated that the altered bacterial community is causal for leaf tissue damage phenotypes. Pairwise bacteria-bacteria competition assay in vitro and in vivo suggested that some Proteobacteria can outcompete most Firmicutes when they are in close proximity. Thus, in the mfec mutant, an increased abundance of leaf endophytic bacteria may stimulate microbe-microbe interactions, contributing to the conversion of a Firmicutes-rich community to a Proteobacteria-rich community. Finally, the authors found a point mutation in CONSTITUTIVELY ACTIVATED CELL DEATH1 (CAD1) gene alone phenocopied the mfec mutant, raising a possibility that CAD1 is a convergent component downstream of pattern-triggered immunity and the MIN7 vesicle-trafficking pathway. The results shown in this study resemble animal dysbiosis, a condition where altered microbiota negatively impacts the host health. This study reveals that the plant pattern-triggered immunity and MIN7 vesicle trafficking pathways have non-redundant and essential roles in preventing dysbiosis and provides insights into underlying molecular mechanisms. (Summary by Tatsuya Nobori @nobolly) Nature 10.1038/s41586-020-2185-0
The large apoplastic intercellular space of plant leaves creates nutrient-rich niches for microbial colonization. To date, whether and how plants control the composition of leaf microbiota is poorly understood. Chen et al. reported that the Arabidopsis quadruple mutant (min7fls2efrcerk1 or mfec) defective in pattern-triggered immunity and a vesicle trafficking pathway (MIN7 encodes an ARF-GEF protein) showed tissue damage phenotypes when grown in soils. The mutant plants showed marked changes in the leaf endophytic bacterial community: 1) an overall increased bacterial population size, 2) decreased community diversity, and 3) a shift from a Firmicutes-rich community to a Proteobacteria-rich community. The authors demonstrated that the altered bacterial community is causal for leaf tissue damage phenotypes. Pairwise bacteria-bacteria competition assay in vitro and in vivo suggested that some Proteobacteria can outcompete most Firmicutes when they are in close proximity. Thus, in the mfec mutant, an increased abundance of leaf endophytic bacteria may stimulate microbe-microbe interactions, contributing to the conversion of a Firmicutes-rich community to a Proteobacteria-rich community. Finally, the authors found a point mutation in CONSTITUTIVELY ACTIVATED CELL DEATH1 (CAD1) gene alone phenocopied the mfec mutant, raising a possibility that CAD1 is a convergent component downstream of pattern-triggered immunity and the MIN7 vesicle-trafficking pathway. The results shown in this study resemble animal dysbiosis, a condition where altered microbiota negatively impacts the host health. This study reveals that the plant pattern-triggered immunity and MIN7 vesicle trafficking pathways have non-redundant and essential roles in preventing dysbiosis and provides insights into underlying molecular mechanisms. (Summary by Tatsuya Nobori @nobolly) Nature 10.1038/s41586-020-2185-0
Extensive inter-plant protein transfer between Cuscuta parasites and their host plants
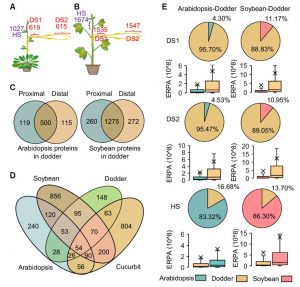 Cuscuta (dodders) are parasites that survive on other host plants.Liu et al. found more than 1500 proteins are transferred between Cucuta and host plants forming a sort of interplant chemical communication. Furthermore, proteins could move between two hosts that are connected by dodder bridges, and retain their activity in the foreign plants. These transferred proteins can be as much as 10% of the proteome of the foreign plant. This work also reveals extensive transfer of protein among cells, tissues and organs, which may be a requirement for regulating the growth and development of the plant in response to environmental conditions. (Summary by Arun Shankar @arunshanker) Mol. Plant 10.1016/j.molp.2019.12.002
Cuscuta (dodders) are parasites that survive on other host plants.Liu et al. found more than 1500 proteins are transferred between Cucuta and host plants forming a sort of interplant chemical communication. Furthermore, proteins could move between two hosts that are connected by dodder bridges, and retain their activity in the foreign plants. These transferred proteins can be as much as 10% of the proteome of the foreign plant. This work also reveals extensive transfer of protein among cells, tissues and organs, which may be a requirement for regulating the growth and development of the plant in response to environmental conditions. (Summary by Arun Shankar @arunshanker) Mol. Plant 10.1016/j.molp.2019.12.002
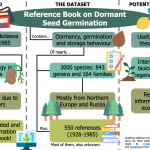
Nikolaeva et al.’s reference book on seed dormancy and germination ($)
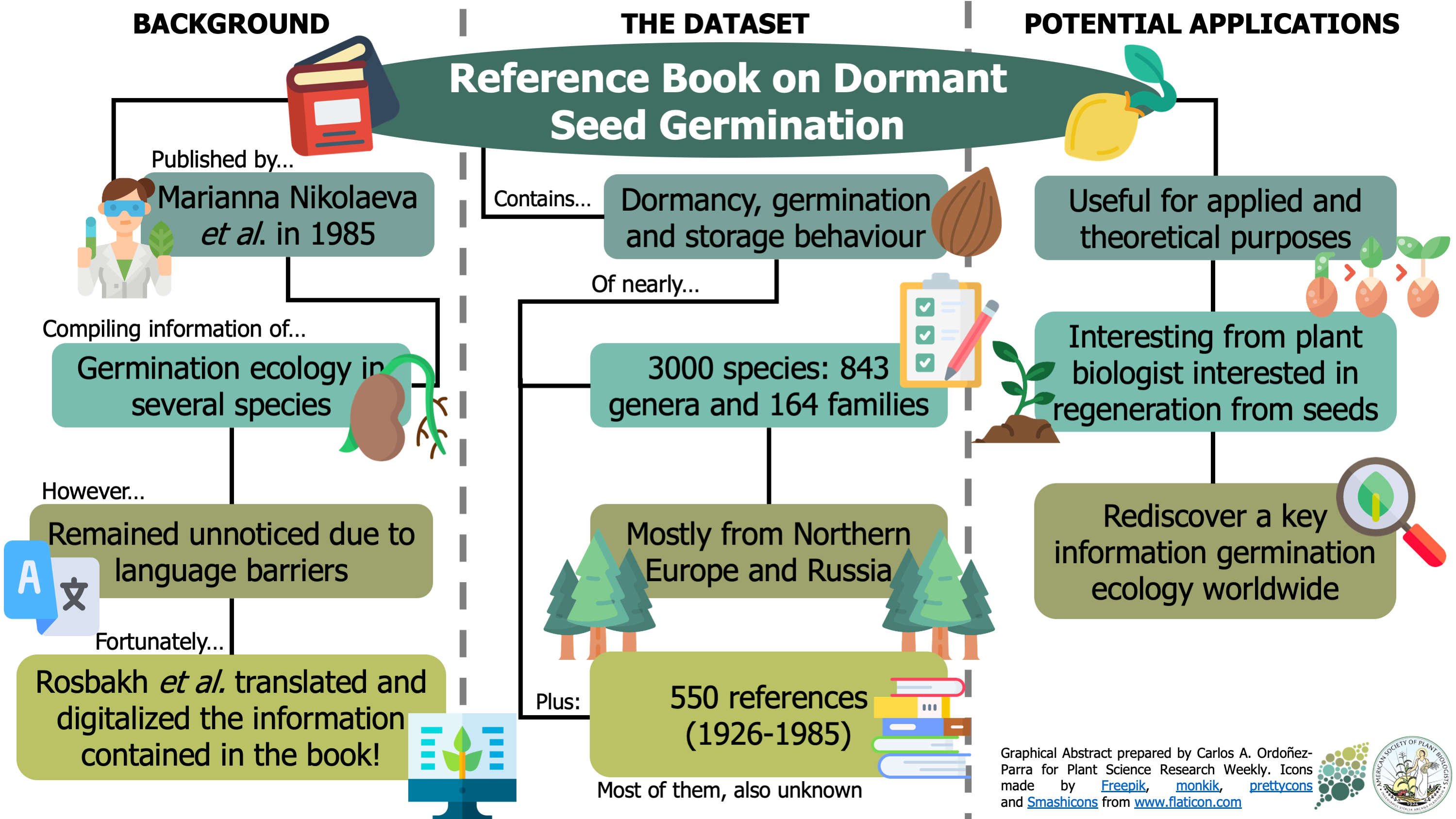 In 1985, Marianna Nikolaeva, Marina Razumova, and Valentina Gladkova published the Reference Book on Dormant Seed Germination. There they provided details on the germination ecology of different plant species –mostly from Northern Europe and Russia. However, since the original publication was in Russian, it remained unnoticed for most seed scientists outside Russia. Fortunately, Rosbakh et al. translated and digitalized the information contained in Nikolaeva et al. publication, and we can now easily access to this vast compendium. This highly valuable dataset –attached to the article as supplemental data– gathers information about the dormancy, germination, and storage behavior of almost 3000 species of seed plants (~1% of the total plant species worldwide). Moreover, it also recovers the nearly 550 seed ecology studies cited in the book published in between 1926-1985. As a result, this data paper allows the seed science community to rediscover a source of information that provides meaningful insights into the development of seed germination ecology worldwide. (Summary by Carlos A. Ordóñez-Parra @caordonezparra Ecology 10.1002/ecy.3049
In 1985, Marianna Nikolaeva, Marina Razumova, and Valentina Gladkova published the Reference Book on Dormant Seed Germination. There they provided details on the germination ecology of different plant species –mostly from Northern Europe and Russia. However, since the original publication was in Russian, it remained unnoticed for most seed scientists outside Russia. Fortunately, Rosbakh et al. translated and digitalized the information contained in Nikolaeva et al. publication, and we can now easily access to this vast compendium. This highly valuable dataset –attached to the article as supplemental data– gathers information about the dormancy, germination, and storage behavior of almost 3000 species of seed plants (~1% of the total plant species worldwide). Moreover, it also recovers the nearly 550 seed ecology studies cited in the book published in between 1926-1985. As a result, this data paper allows the seed science community to rediscover a source of information that provides meaningful insights into the development of seed germination ecology worldwide. (Summary by Carlos A. Ordóñez-Parra @caordonezparra Ecology 10.1002/ecy.3049