Plant Science Research Weekly: April 2, 2021
Update: Lateral root formation and nutrients: nitrogen in the spotlight
 We marvel at the ability of starfish to regenerate an arm, or a lizard a new tail, but even more impressive than these feats is the ability of a plant root system to create lateral roots when and where they are needed. This root system plasticity has been long studied, with a wealth of insights into the molecular mechanisms of both lateral root formation and nutrient sensing. Here, Pélissier, Mott and Bleeckman summarize recent findings on the intersection of these two fields, with an emphasis on the role of nitrogen in lateral root formation. As the authors describe, nitrate and its sister nutrient ammonium play key roles at several stages of lateral root formation, from cell divisions in the progenitor pericycle cells to emergence. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab145
We marvel at the ability of starfish to regenerate an arm, or a lizard a new tail, but even more impressive than these feats is the ability of a plant root system to create lateral roots when and where they are needed. This root system plasticity has been long studied, with a wealth of insights into the molecular mechanisms of both lateral root formation and nutrient sensing. Here, Pélissier, Mott and Bleeckman summarize recent findings on the intersection of these two fields, with an emphasis on the role of nitrogen in lateral root formation. As the authors describe, nitrate and its sister nutrient ammonium play key roles at several stages of lateral root formation, from cell divisions in the progenitor pericycle cells to emergence. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiab145
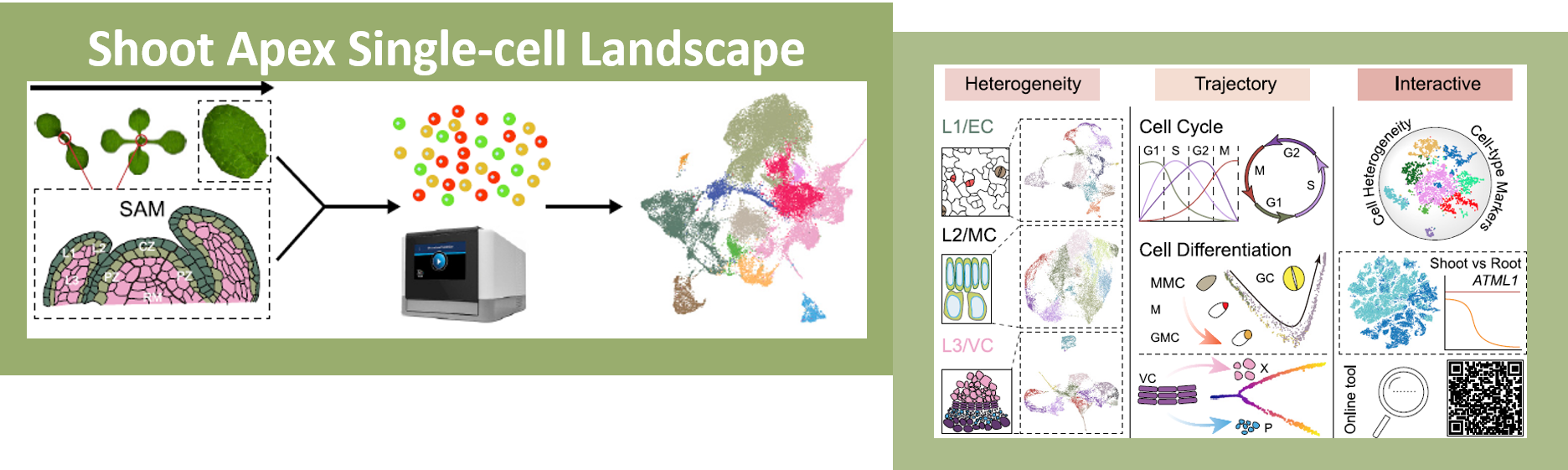
A single-cell analysis of the Arabidopsis vegetative shoot apex
One of the most fascinating challenges in developmental biology is to understand how a few stem cells hidden in a microscopic region of the shoot apex give origin to all aerial parts of the plant. Technological advances such as single-cell RNA-sequencing (scRNA-seq) are enabling the comprehensive analysis of heterogeneous cell populations with unprecedent resolution. Zhang and coworkers previously employed scRNA-seq to determine the transcriptome profiles of the root apex in Arabidopsis. Here, they investigate stem cell functioning and cell fate determination of the shoot apex, and identified 23 cell clusters visualized into 7 broad populations. Transcriptome analysis of the shoot apex revealed accumulation of leaf polarity genes in a group of meristematic cells, indicating that these transit-amplification cells (i.e., in transition from undifferentiated to differentiated conditions) displayed a continuum of expression states during leaf primordia initiation. The authors also reconstructed developmental trajectories of different tissues within the shoot apex by combining spatial distribution and temporal ordering of individual cells, and arranged clusters in cyclic patterns depending on the transcript levels of core cell-cycle genes. Sequencing the transcriptomes of individual cells represents a revolutionary approach to decipher plant development and to accelerate the discovery of novel genes, with the resulting Root and Shoot Cell Atlases constituting valuable resources for the plant science community. You can search for your favorite genes at http://wanglab.sippe.ac.cn/shootatlas/. (Summary and image adaptation by Michela Osnato @michela_osnato) Dev. Cell. 10.1016/j.devcel.2021.02.021
A clustered mitochondria family protein mediates the plant mitophagy
 Mitochondria function as cellular powerhouses to generate energy via oxidative phosphorylation and facilitate the synthesis of essential macromolecules. To protect against proteotoxic stress, damaged mitochondria are selectively removed by autophagy via a process known as mitophagy. In mammalian cells, the selective mitophagy receptors and adaptors proteins have been extensively studied, but those homologs are mostly absent from the plant genome. Previusly, an Arabidopsis mutant named friendly was identified in which mitochondria cluster together; FRIENDLY is a member of the conserved CLUSTERED MITOCHONDRIA family. To study the process of mitophagy, Ma et al. used uncouplers of oxidative phosphorylation such as the proton ionophore DNP to induce mitophagy. Depolarized mitochondria are selectively engulfed by autophagosomes, dependent on ATG5. Uncoupler treatments also induce ubiquitination of the outer mitochondria membrane, involving both mitophagy and proteasome degradation pathways. The authors found the friendly mutant has less mitophagosomes and displays defective elimination of damaged mitochondria, indicating that FRIENDLY is essential for uncoupler-induced mitophagy. Interestingly, the induced mitophagy causes delay of greening cotyledon in the friendly and atg5 mutants, suggesting its role in metabolic switching. This study sheds lights on how plants employ mitophagy in inter-organelle communication under stress and development. (Summary by Min May Wong @wongminmay) Curr. Biol. 10.1016/j.cub.2021.02.034
Mitochondria function as cellular powerhouses to generate energy via oxidative phosphorylation and facilitate the synthesis of essential macromolecules. To protect against proteotoxic stress, damaged mitochondria are selectively removed by autophagy via a process known as mitophagy. In mammalian cells, the selective mitophagy receptors and adaptors proteins have been extensively studied, but those homologs are mostly absent from the plant genome. Previusly, an Arabidopsis mutant named friendly was identified in which mitochondria cluster together; FRIENDLY is a member of the conserved CLUSTERED MITOCHONDRIA family. To study the process of mitophagy, Ma et al. used uncouplers of oxidative phosphorylation such as the proton ionophore DNP to induce mitophagy. Depolarized mitochondria are selectively engulfed by autophagosomes, dependent on ATG5. Uncoupler treatments also induce ubiquitination of the outer mitochondria membrane, involving both mitophagy and proteasome degradation pathways. The authors found the friendly mutant has less mitophagosomes and displays defective elimination of damaged mitochondria, indicating that FRIENDLY is essential for uncoupler-induced mitophagy. Interestingly, the induced mitophagy causes delay of greening cotyledon in the friendly and atg5 mutants, suggesting its role in metabolic switching. This study sheds lights on how plants employ mitophagy in inter-organelle communication under stress and development. (Summary by Min May Wong @wongminmay) Curr. Biol. 10.1016/j.cub.2021.02.034
Whitefly hijacks a plant detoxification gene that neutralizes plant toxins
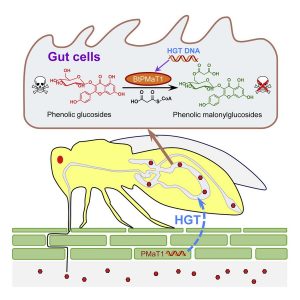 The sweetpotato whitefly, Bemisia tabaci, are devastating crop pests that lower yields and transmit viruses. They feed off more than 600 species, most of which produce toxic phenolic glycosides. Xia et al. investigated how the whiteflies manage to avoid the effects of the toxin. They found convincing evidence that this group of insects, but not their closest relatives, carry a gene encoding a BAHD acyltransferases, a class of enzymes known to detoxify phenolic glycosides. Remarkably, the nearest relatives to this insect gene are plant genes encoding BAHD acyltransferases, indicating that the insects acquired the gene from the plants they feed on. The authors showed that silencing this gene in the insects led to increased mortality upon feeding, and also demonstrated that the encoded enzyme has the detoxifying function in vitro and in vivo. Introducing dsRNA targeting the insect gene into tomato leaves enhanced resistance to the whiteflies. This study thus shows that horizontal gene transfer of a plant gene has enabled the whiteflies to overcome the plant’s own defense mechanism, but witih a little help the plant can regain the upper hand in this arms race. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2021.02.014
The sweetpotato whitefly, Bemisia tabaci, are devastating crop pests that lower yields and transmit viruses. They feed off more than 600 species, most of which produce toxic phenolic glycosides. Xia et al. investigated how the whiteflies manage to avoid the effects of the toxin. They found convincing evidence that this group of insects, but not their closest relatives, carry a gene encoding a BAHD acyltransferases, a class of enzymes known to detoxify phenolic glycosides. Remarkably, the nearest relatives to this insect gene are plant genes encoding BAHD acyltransferases, indicating that the insects acquired the gene from the plants they feed on. The authors showed that silencing this gene in the insects led to increased mortality upon feeding, and also demonstrated that the encoded enzyme has the detoxifying function in vitro and in vivo. Introducing dsRNA targeting the insect gene into tomato leaves enhanced resistance to the whiteflies. This study thus shows that horizontal gene transfer of a plant gene has enabled the whiteflies to overcome the plant’s own defense mechanism, but witih a little help the plant can regain the upper hand in this arms race. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2021.02.014
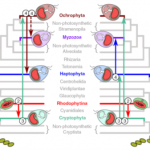
A molecular timescale for eukaryote evolution with implications for the origin of red algal-derived plastids
 Algae powered by red algal-derived plastids (e.g., diatoms, dinoflagellates, and coccolithophores) are among the most evolutionary and ecologically successful eukaryotes on Earth, but their origins and relationships remain poorly understood. Here, Strassert et al. test the rhodoplex hypothesis that designates an evolutionary scenario in which plastids were acquired from red algae by a single secondary endosymbiosis, with further horizontal spread in other eukaryotes by eukaryote-to-eukaryote endosymbiosis. The study shows that the recipients and the donors of plastids were contemporaneous and most likely overlapped during the early history of the life on Earth, which explains transfer of these plastids between seemingly unlikely distant related hosts. The study also shows that the time between the start of the secondary endosymbiosis and the establishment of the complex red plastids was in a short time frame between 650 to 1079 million years. Interestingly, in spite of their ancient origins, red algal-derived plastid-containing lineages did not become ecological prominent for another billion years, perhaps coinciding with the breakup of continents and formation of nutrient-rich coastal zones. (Summary by Arun K. Shanker @arunshanker) Nature Comms. 10.1038/s41467-021-22044-z
Algae powered by red algal-derived plastids (e.g., diatoms, dinoflagellates, and coccolithophores) are among the most evolutionary and ecologically successful eukaryotes on Earth, but their origins and relationships remain poorly understood. Here, Strassert et al. test the rhodoplex hypothesis that designates an evolutionary scenario in which plastids were acquired from red algae by a single secondary endosymbiosis, with further horizontal spread in other eukaryotes by eukaryote-to-eukaryote endosymbiosis. The study shows that the recipients and the donors of plastids were contemporaneous and most likely overlapped during the early history of the life on Earth, which explains transfer of these plastids between seemingly unlikely distant related hosts. The study also shows that the time between the start of the secondary endosymbiosis and the establishment of the complex red plastids was in a short time frame between 650 to 1079 million years. Interestingly, in spite of their ancient origins, red algal-derived plastid-containing lineages did not become ecological prominent for another billion years, perhaps coinciding with the breakup of continents and formation of nutrient-rich coastal zones. (Summary by Arun K. Shanker @arunshanker) Nature Comms. 10.1038/s41467-021-22044-z