Plant Science Research Weekly: April 10
White paper: Introducing plant biology graduate students to a culture of mental well‐being
 Graduate studies are challenging times, fraught with uncertainty and stress. Dewa et al. addressed this problem head on by developing a required, credited course for first-year graduate students, “Tools for Becoming a Successful Professional and for Enhancing Your Well‐Being and Work Environment”. The five-week course included discussions about emotion-focused and problem-focused coping, as well as evaluations of the scientific literature supporting various strategies. The fact that the course was required and given for credit signaled to the students that the graduate program takes their mental health seriously. The authors also note that besides providing graduate students with the opportunity to reflect on their own approach to mental well-being, it also provided them with tools to share with the undergraduates they would encounter in their roles as teaching assistants. (Summary by Mary Williams) Plant Direct 10.1002/pld3.211
Graduate studies are challenging times, fraught with uncertainty and stress. Dewa et al. addressed this problem head on by developing a required, credited course for first-year graduate students, “Tools for Becoming a Successful Professional and for Enhancing Your Well‐Being and Work Environment”. The five-week course included discussions about emotion-focused and problem-focused coping, as well as evaluations of the scientific literature supporting various strategies. The fact that the course was required and given for credit signaled to the students that the graduate program takes their mental health seriously. The authors also note that besides providing graduate students with the opportunity to reflect on their own approach to mental well-being, it also provided them with tools to share with the undergraduates they would encounter in their roles as teaching assistants. (Summary by Mary Williams) Plant Direct 10.1002/pld3.211
Perspective: What is replication?
 This is an interesting paper and certainly one to share with students. Nosek and Errington argue that how we usually think of as “replication”, repeating a study and observing the same results, is incorrect. Instead, they argue for a broader definition of replication, as something that supports the findings, irrespective of the methods used. The authors argue that successful replication indicates that the findings are generalizable, whereas unsuccessful efforts indicate that the original finding might be constrained. To put it another way, ”outcomes consistent with a prior claim would increase confidence in the claim, and outcomes inconsistent with a prior claim would decrease confidence in the claim.” The authors suggest that redefining replication should remove some of the stigma that makes authors reluctant to pursue it, journals reluctant to publish it, and funders reluctant to fund it. This is a thought-provoking paper, and also includes a fascinating anecdote of summer versus winter frogs – have a look! (Summary by Mary Williams) PLOS Biol. 10.1371/journal.pbio.3000691
This is an interesting paper and certainly one to share with students. Nosek and Errington argue that how we usually think of as “replication”, repeating a study and observing the same results, is incorrect. Instead, they argue for a broader definition of replication, as something that supports the findings, irrespective of the methods used. The authors argue that successful replication indicates that the findings are generalizable, whereas unsuccessful efforts indicate that the original finding might be constrained. To put it another way, ”outcomes consistent with a prior claim would increase confidence in the claim, and outcomes inconsistent with a prior claim would decrease confidence in the claim.” The authors suggest that redefining replication should remove some of the stigma that makes authors reluctant to pursue it, journals reluctant to publish it, and funders reluctant to fund it. This is a thought-provoking paper, and also includes a fascinating anecdote of summer versus winter frogs – have a look! (Summary by Mary Williams) PLOS Biol. 10.1371/journal.pbio.3000691
Review: The genetic basis and nutritional benefits of pigmented rice
 Pigmented rice varieties are those in which a pigment is deposited in the bran, the outer layer of the grain. The pigment can be from brown to red (proanthocyanidins) or from purple to black (anthocyanins). In the course of domestication, humans have selected against the genetic factors responsible for pigmentation due to their linkage with traits controlling grain shattering and seed dormancy. However, indigenous knowledge of some rice-consuming populations suggests that pigmented rice possesses a plethora of health-befitting properties such as anti-inflammatory, anti-diabetic, anti-hyperlipidemic, anti-arthritic, anti-cancerous and diuretic activities; modern scientific evidence sometimes also supports the traditional knowledge. Mbanjo et al. review the nutritional and health benefits of pigmented rice. They discuss the phytochemicals behind the benefits, the genetic factors and mechanisms behind production of the phytochemicals, and how these factors and mechanisms are identified. They conclude with a discussion of efforts to develop pigmented rice varieties to meet the rising demand for their consumption. (Summary by Hari Gowthem G) Frontiers in Genetics 10.3389/fgene.2020.00229
Pigmented rice varieties are those in which a pigment is deposited in the bran, the outer layer of the grain. The pigment can be from brown to red (proanthocyanidins) or from purple to black (anthocyanins). In the course of domestication, humans have selected against the genetic factors responsible for pigmentation due to their linkage with traits controlling grain shattering and seed dormancy. However, indigenous knowledge of some rice-consuming populations suggests that pigmented rice possesses a plethora of health-befitting properties such as anti-inflammatory, anti-diabetic, anti-hyperlipidemic, anti-arthritic, anti-cancerous and diuretic activities; modern scientific evidence sometimes also supports the traditional knowledge. Mbanjo et al. review the nutritional and health benefits of pigmented rice. They discuss the phytochemicals behind the benefits, the genetic factors and mechanisms behind production of the phytochemicals, and how these factors and mechanisms are identified. They conclude with a discussion of efforts to develop pigmented rice varieties to meet the rising demand for their consumption. (Summary by Hari Gowthem G) Frontiers in Genetics 10.3389/fgene.2020.00229
Genome-phenome wide association in maize and Arabidopsis identifies a common molecular and evolutionary signature
 Genome-wide association studies (GWAS) are widely used to link natural genetic variation to trait variation, with a single or a select few correlated traits assessed. High-throughput phenotyping allows the scoring of hundreds of individuals for various traits at several time points. An undeniable consequence of quantitative genetics research is the creation of large, complex datasets that can contain information on a multitude of traits in a population. Here, Liang et al. conducted a Genome-Phenome Wide Association Study (GPWAS) on maize and Arabidopsis. While a conventional GWAS only considers traits that are correlated, this novel GPWAS approach links gene markers to several correlated and uncorrelated traits simultaneously. The authors found that genes identified using this “multi-trait and multi-SNP framework” are more similar to those previously characterized using forward genetics approaches than genes from annotated gene models or conventional GWAS. GPWAS-identified maize genes resemble those with verified loss-of-function phenotypes at the molecular, population, and evolutionary levels by higher expression levels, syntenic conservation, and stronger purifying selection respectively, with similar observations made for Arabidopsis. Interestingly, genes linked to variation in individual phenotypes by GWAS had intermediate values between the annotated gene models and GPWAS/classical mutants. This may suggest that GPWAS is more likely to detect pleiotropic genes while conventional GWAS is more suitable for identifying loci that control a smaller number of phenotypes. Nonetheless, GPWAS represents an interesting approach for using the wealth of data available for model organisms such as maize, rice, and Arabidopsis. (Summary by Caroline Dowling) Mol. Plant 10.1016/j.molp.2020.03.003
Genome-wide association studies (GWAS) are widely used to link natural genetic variation to trait variation, with a single or a select few correlated traits assessed. High-throughput phenotyping allows the scoring of hundreds of individuals for various traits at several time points. An undeniable consequence of quantitative genetics research is the creation of large, complex datasets that can contain information on a multitude of traits in a population. Here, Liang et al. conducted a Genome-Phenome Wide Association Study (GPWAS) on maize and Arabidopsis. While a conventional GWAS only considers traits that are correlated, this novel GPWAS approach links gene markers to several correlated and uncorrelated traits simultaneously. The authors found that genes identified using this “multi-trait and multi-SNP framework” are more similar to those previously characterized using forward genetics approaches than genes from annotated gene models or conventional GWAS. GPWAS-identified maize genes resemble those with verified loss-of-function phenotypes at the molecular, population, and evolutionary levels by higher expression levels, syntenic conservation, and stronger purifying selection respectively, with similar observations made for Arabidopsis. Interestingly, genes linked to variation in individual phenotypes by GWAS had intermediate values between the annotated gene models and GPWAS/classical mutants. This may suggest that GPWAS is more likely to detect pleiotropic genes while conventional GWAS is more suitable for identifying loci that control a smaller number of phenotypes. Nonetheless, GPWAS represents an interesting approach for using the wealth of data available for model organisms such as maize, rice, and Arabidopsis. (Summary by Caroline Dowling) Mol. Plant 10.1016/j.molp.2020.03.003
Versatile auto-luminescent luciferase-based reporters for spatiotemporal gene expression studies
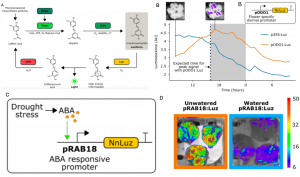 Firefly luciferase constructs are extensively used for gene expression studies but require a reaction with an exogenous substrate, so expression to some extent reflects tissue penetrance. Khakhar et al. designed customizable auto-luminescence constructs based on a fungal bioluminescence pathway (FBP) in which caffeic acid, an intermediate of lignin biosynthesis, is converted into luciferin by the action of three fungal enzymes. Auto-luminescence was observed following infiltration with Agrobacterium carrying the FBP constructs into Nicotiana leaves, with the luminescence enhanced by also introducing an enzyme that recycles the byproduct of the luciferase reaction or engineering increased expression of caffeic acid biosynthesis. These constructs can be modified to use in a variety of plant species like Arabidopsis, petunia, tomato, dahlia, periwinkle, and rose. As examples, using this system the authors observed a diurnal expression pattern from the ODORANT1 promoter in petunia and changes in transcription in response to abscisic acid flux in Arabidopsis. Other advantages are that the signal of the luciferase reaction is emitted in the green spectrum, making it compatible to use along with the conventional firefly luciferase assay, and imaging uses an entry-level DSLR camera along with Raspberry Pi single board computer instead of more expensive CCD cameras. These new tools offer an economic way to study gene expression changes over long periods during environmental and hormonal changes and can be used to study plant-pollinator interactions and many other creative ways. (Summary by Vijaya Batthula) eLife 10.7554/eLife.52786
Firefly luciferase constructs are extensively used for gene expression studies but require a reaction with an exogenous substrate, so expression to some extent reflects tissue penetrance. Khakhar et al. designed customizable auto-luminescence constructs based on a fungal bioluminescence pathway (FBP) in which caffeic acid, an intermediate of lignin biosynthesis, is converted into luciferin by the action of three fungal enzymes. Auto-luminescence was observed following infiltration with Agrobacterium carrying the FBP constructs into Nicotiana leaves, with the luminescence enhanced by also introducing an enzyme that recycles the byproduct of the luciferase reaction or engineering increased expression of caffeic acid biosynthesis. These constructs can be modified to use in a variety of plant species like Arabidopsis, petunia, tomato, dahlia, periwinkle, and rose. As examples, using this system the authors observed a diurnal expression pattern from the ODORANT1 promoter in petunia and changes in transcription in response to abscisic acid flux in Arabidopsis. Other advantages are that the signal of the luciferase reaction is emitted in the green spectrum, making it compatible to use along with the conventional firefly luciferase assay, and imaging uses an entry-level DSLR camera along with Raspberry Pi single board computer instead of more expensive CCD cameras. These new tools offer an economic way to study gene expression changes over long periods during environmental and hormonal changes and can be used to study plant-pollinator interactions and many other creative ways. (Summary by Vijaya Batthula) eLife 10.7554/eLife.52786
ORHis, a natural variant of OR, interacts with plastid division factor ARC3 to regulate chromoplast number and carotenoid accumulation
 Some “superfoods” have high nutritional value due to the presence of carotenoids, which prevent degenerative diseases like cancer. In plants, these pigments are biosynthesized and stored by plastid organelles called chromoplasts. Chromoplast number and size define total carotenoid accumulation. ORHis, a natural allele of Orange (OR) in which a conserved Arg is replaced with a His, both promotes carotenoid accumulation in melon fruit, and decreases the number of chromoplasts per cell. Sun et al. investigated an ORHis variant of Arabidopsis OR and confirmed its effects as a chromoplast number regulator in addition to its functions in carotenoid biosynthesis and chromoplast biogenesis. The authors discovered that ORHis constrains chromoplast number by interacting with ARC3 (a central chloroplast division factor) and suppressing its interaction with PARC6 (another key regulator of chloroplast division). Additionally, the manipulation of the plastid division factors ARC3 and PDV1 significantly affected carotenoid levels in Arabidopsis ORHis calli, which provides a novel strategy for carotenoid enrichment in food crops. (Summary by Elisandra Pradella) Mol. Plant 10.1016/j.molp.2020.03.007
Some “superfoods” have high nutritional value due to the presence of carotenoids, which prevent degenerative diseases like cancer. In plants, these pigments are biosynthesized and stored by plastid organelles called chromoplasts. Chromoplast number and size define total carotenoid accumulation. ORHis, a natural allele of Orange (OR) in which a conserved Arg is replaced with a His, both promotes carotenoid accumulation in melon fruit, and decreases the number of chromoplasts per cell. Sun et al. investigated an ORHis variant of Arabidopsis OR and confirmed its effects as a chromoplast number regulator in addition to its functions in carotenoid biosynthesis and chromoplast biogenesis. The authors discovered that ORHis constrains chromoplast number by interacting with ARC3 (a central chloroplast division factor) and suppressing its interaction with PARC6 (another key regulator of chloroplast division). Additionally, the manipulation of the plastid division factors ARC3 and PDV1 significantly affected carotenoid levels in Arabidopsis ORHis calli, which provides a novel strategy for carotenoid enrichment in food crops. (Summary by Elisandra Pradella) Mol. Plant 10.1016/j.molp.2020.03.007
Structural basis for WUSCHEL binding
 The transcription factor WUSCHEL (WUS) plays a central role in organization of the shoot meristem. The three-helix bundle homeodomain in WUS can bind to several distinct DNA sequence motifs in many target genes promoters, but a structural view of these binding events has been lacking. Here Sloan et al. solve the structure of the WUS homeodomain (WUS-HD) bound to DNA probes containing “TAAT”, tandem “TGAA” repeats, and the G-box sequences derived from WUS-regulated genes. Overall, the WUS-HD interacts with DNA as a typical homeodomain, with conserved residues in the recognition helix inserting into the DNA major groove, but the authors also find non-conserved, WUS family-specific residues that make significant protein-DNA contacts. In contrast to many other homeodomain transcription factors, in all cases WUS most likely binds the target as a dimer. The authors then provide evidence, including cooperative binding through DNA-mediated WUS-WUS interactions and a favorable DNA helix shape, for why the direct TGAATGAA repeat is the most high-affinity target, followed by the G-box and TAAT. By comparing their results to whole genome ChiP-seq data, they further suggest that these relative binding affinities obtained in vitro actually translate to real cells. Altogether, these results provide a rational foundation for further exploration of the complex regulatory landscape of WUS-dependent transcription. (Summary by Frej Tulin) bioRxiv 10.1101/2020.03.26.009761v2
The transcription factor WUSCHEL (WUS) plays a central role in organization of the shoot meristem. The three-helix bundle homeodomain in WUS can bind to several distinct DNA sequence motifs in many target genes promoters, but a structural view of these binding events has been lacking. Here Sloan et al. solve the structure of the WUS homeodomain (WUS-HD) bound to DNA probes containing “TAAT”, tandem “TGAA” repeats, and the G-box sequences derived from WUS-regulated genes. Overall, the WUS-HD interacts with DNA as a typical homeodomain, with conserved residues in the recognition helix inserting into the DNA major groove, but the authors also find non-conserved, WUS family-specific residues that make significant protein-DNA contacts. In contrast to many other homeodomain transcription factors, in all cases WUS most likely binds the target as a dimer. The authors then provide evidence, including cooperative binding through DNA-mediated WUS-WUS interactions and a favorable DNA helix shape, for why the direct TGAATGAA repeat is the most high-affinity target, followed by the G-box and TAAT. By comparing their results to whole genome ChiP-seq data, they further suggest that these relative binding affinities obtained in vitro actually translate to real cells. Altogether, these results provide a rational foundation for further exploration of the complex regulatory landscape of WUS-dependent transcription. (Summary by Frej Tulin) bioRxiv 10.1101/2020.03.26.009761v2
Deubiquitinating enzymes UBP12, 13 are antagonistic to E3 ligases BIG BROTHER and DA2
 Post-translation modifications like ubiquitination and deubiquitination are important to maintain protein homeostasis and in turn plant growth. DA1, and DA1-RELATED PROTEIN 1, 2 (DAR1, DAR2) are latent peptidases activated upon ubiquitination by E3 ligases BIG BROTHER (BB) and DA2. In this paper, Vanhaeren et al. identified UBIQUITIN BINDING PROTEIN 12, 13 (UBP12, 13) as the interacting partners of DA1, DAR1, and DAR2 in vitro and in vivo. Overexpression of UBP12, 13 decreases rosette and leaf size, due to decreased cell size because of delayed onset of cellular differentiation and endoreduplication. Decreased leaf size was also observed when both UBP12, 13 were downregulated. In vitro and in vivo experiments showed that UBP12, 13 interact and deubiquitinate DA1, DAR1, DAR2 rendering the active peptidases inactive. Molecular and co-localization studies showed that UBP12, 13 and DA1, DAR1, DAR2 co-express and co-localize throughout the leaf development. Further, UBP12 and 13 are not targeted by DA1 as opposed to BB, DA2, UBP15 which are targeted by DA1. Thus UBP12, 13 acts upstream of DA1, DAR1, DAR2 in regulating plant growth. (Summary by Vijaya Batthula) eLIFE 10.7554/eLife.52276
Post-translation modifications like ubiquitination and deubiquitination are important to maintain protein homeostasis and in turn plant growth. DA1, and DA1-RELATED PROTEIN 1, 2 (DAR1, DAR2) are latent peptidases activated upon ubiquitination by E3 ligases BIG BROTHER (BB) and DA2. In this paper, Vanhaeren et al. identified UBIQUITIN BINDING PROTEIN 12, 13 (UBP12, 13) as the interacting partners of DA1, DAR1, and DAR2 in vitro and in vivo. Overexpression of UBP12, 13 decreases rosette and leaf size, due to decreased cell size because of delayed onset of cellular differentiation and endoreduplication. Decreased leaf size was also observed when both UBP12, 13 were downregulated. In vitro and in vivo experiments showed that UBP12, 13 interact and deubiquitinate DA1, DAR1, DAR2 rendering the active peptidases inactive. Molecular and co-localization studies showed that UBP12, 13 and DA1, DAR1, DAR2 co-express and co-localize throughout the leaf development. Further, UBP12 and 13 are not targeted by DA1 as opposed to BB, DA2, UBP15 which are targeted by DA1. Thus UBP12, 13 acts upstream of DA1, DAR1, DAR2 in regulating plant growth. (Summary by Vijaya Batthula) eLIFE 10.7554/eLife.52276
Newly identified miRNAs may contribute to aerenchyma formation in sugarcane roots
 Sugarcane is one of the major crops for sugar and ethanol production and has been successfully used for first generation ethanol production through sucrose fermentation, but second generation ethanol production, through cell wall depolymerization, is still limited. Aerenchyma formation in sugarcane root occurs through the digestion of cell walls but the process controlling it remains unclear. In this paper, Traves et al. show potential epigenetic control during aerenchyma formation through the identification of 39 miRNAs in sugarcane. Gene ontology enrichment analysis of putative transcript targets of these miRNAs revealed that 2.3 and 3.5% of the assigned targets were enriched in cell wall organization and carbohydrate metabolism categories respectively. Considering various modules involved in the formation of aerenchyma, the identified miRNA targets seem to be involved in hormone perception, cell separation, and hemicellulose hydrolysis modules. Identification of 13 miRNAs potentially involved in ethylene perception and signaling supports previous findings on the role of ethylene during aerenchyma formation. Further study of identified miRNAs should provide a deeper understanding of cell wall degradation and hydrolysis of cell wall polymers and contribute to the production of biofuels. (Summary by Sunita Pathak) Plant Direct 10.1002/pld3.204
Sugarcane is one of the major crops for sugar and ethanol production and has been successfully used for first generation ethanol production through sucrose fermentation, but second generation ethanol production, through cell wall depolymerization, is still limited. Aerenchyma formation in sugarcane root occurs through the digestion of cell walls but the process controlling it remains unclear. In this paper, Traves et al. show potential epigenetic control during aerenchyma formation through the identification of 39 miRNAs in sugarcane. Gene ontology enrichment analysis of putative transcript targets of these miRNAs revealed that 2.3 and 3.5% of the assigned targets were enriched in cell wall organization and carbohydrate metabolism categories respectively. Considering various modules involved in the formation of aerenchyma, the identified miRNA targets seem to be involved in hormone perception, cell separation, and hemicellulose hydrolysis modules. Identification of 13 miRNAs potentially involved in ethylene perception and signaling supports previous findings on the role of ethylene during aerenchyma formation. Further study of identified miRNAs should provide a deeper understanding of cell wall degradation and hydrolysis of cell wall polymers and contribute to the production of biofuels. (Summary by Sunita Pathak) Plant Direct 10.1002/pld3.204
Sex chromosome evolution in asparagus
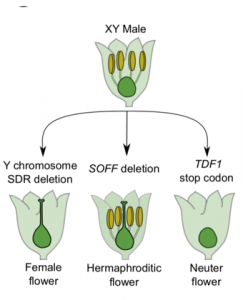 Separation of male and female flowers on different individuals, called dioecy, has evolved independently many times in flowering plants from hermaphroditic ancestors. A long-standing theory predicts that specialized X and Y sex chromosomes can evolve in dioecious species when mutations occur in two tightly linked genes with antagonistic effects on male and female development. In this paper, Harkess et al. provide functional evidence for this two-gene model of sex chromosome evolution in garden asparagus (Asparagus officinalis). In asparagus, the X and Y chromosomes are cytologically homomorphic, but the Y chromosome contains a ~1 Mb, hemizygous sex-determining region. Previous work showed that this sex-determining region contains the gene SUPPRESSOR OF FEMALE FUNCTION (SOFF1) that dominantly blocks pistil development. Here, they extend this analysis by demonstrating the requirement for the closely linked DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION (TDF) in anther development. The combined activities of TOFF1 and TDF1 nicely explain how sex is genetically determined in the dioecious asparagus and gives new insights into plant sex chromosome evolution. (Summary by Frej Tulin) Plant Cell 10.1105/tpc.19.00859
Separation of male and female flowers on different individuals, called dioecy, has evolved independently many times in flowering plants from hermaphroditic ancestors. A long-standing theory predicts that specialized X and Y sex chromosomes can evolve in dioecious species when mutations occur in two tightly linked genes with antagonistic effects on male and female development. In this paper, Harkess et al. provide functional evidence for this two-gene model of sex chromosome evolution in garden asparagus (Asparagus officinalis). In asparagus, the X and Y chromosomes are cytologically homomorphic, but the Y chromosome contains a ~1 Mb, hemizygous sex-determining region. Previous work showed that this sex-determining region contains the gene SUPPRESSOR OF FEMALE FUNCTION (SOFF1) that dominantly blocks pistil development. Here, they extend this analysis by demonstrating the requirement for the closely linked DEFECTIVE IN TAPETAL DEVELOPMENT AND FUNCTION (TDF) in anther development. The combined activities of TOFF1 and TDF1 nicely explain how sex is genetically determined in the dioecious asparagus and gives new insights into plant sex chromosome evolution. (Summary by Frej Tulin) Plant Cell 10.1105/tpc.19.00859
CsIVP functions in vasculature development and downy mildew resistance in cucumber
 High yielding crops are often less resistant to pathogens and vice versa, suggesting that there is an underlying mechanism co-regulating development and disease resistance in plants. Yan et al. identified a transcription factor in cucumber (CsIVP) that regulates vascular development and resistance to downy mildew (caused by the oomycete pathogen Pseudoperonospora cubensis). CsIVP is highly expressed in the vascular tissues and localized in the nucleus. RNAi-mediated knockdown of CsIVP resulted in plants with compromised vascular development and accumulation of auxin in the leaf veins. RNA-seq analysis revealed that several known developmental regulators and auxin-related genes were significantly altered in CsIVP-RNAi compared to the wildtype plants. CsIVP bound E-box elements in the promoters of vascular related genes including CsYAB5. Knockdown of CsYAB5 mimics the phenotype of CsIVP suggesting that CsYAB5 functions downstream of CsIVP in regulating vascular development. CsIVP-RNAi lines were exceptionally resistant to greenhouse diseases thus suggesting the possible role of CsIVP in pathogen response. CsIVP-RNAi plants were specifically resistant to downy mildew, showing reduced lesions and absence of conidiophores compared to WT. Defense-related phytohormone genes were significantly enriched in CsIVP-RNAi. Indeed, salicylic acid (SA) was significantly increased in CsIVP-RNAi leaves. Taken together, CsIVP functions as a regulator of organ morphogenesis and resistance to down mildew through interactions with CsYAB5 and CsNIMIN1 respectively. (Summary by Toluwase Olukayode) PLOS Biol. 10.1371/journal.pbio.3000671
High yielding crops are often less resistant to pathogens and vice versa, suggesting that there is an underlying mechanism co-regulating development and disease resistance in plants. Yan et al. identified a transcription factor in cucumber (CsIVP) that regulates vascular development and resistance to downy mildew (caused by the oomycete pathogen Pseudoperonospora cubensis). CsIVP is highly expressed in the vascular tissues and localized in the nucleus. RNAi-mediated knockdown of CsIVP resulted in plants with compromised vascular development and accumulation of auxin in the leaf veins. RNA-seq analysis revealed that several known developmental regulators and auxin-related genes were significantly altered in CsIVP-RNAi compared to the wildtype plants. CsIVP bound E-box elements in the promoters of vascular related genes including CsYAB5. Knockdown of CsYAB5 mimics the phenotype of CsIVP suggesting that CsYAB5 functions downstream of CsIVP in regulating vascular development. CsIVP-RNAi lines were exceptionally resistant to greenhouse diseases thus suggesting the possible role of CsIVP in pathogen response. CsIVP-RNAi plants were specifically resistant to downy mildew, showing reduced lesions and absence of conidiophores compared to WT. Defense-related phytohormone genes were significantly enriched in CsIVP-RNAi. Indeed, salicylic acid (SA) was significantly increased in CsIVP-RNAi leaves. Taken together, CsIVP functions as a regulator of organ morphogenesis and resistance to down mildew through interactions with CsYAB5 and CsNIMIN1 respectively. (Summary by Toluwase Olukayode) PLOS Biol. 10.1371/journal.pbio.3000671
Auxin response factor targeted by viruses
 Auxin regulates various aspects of plant growth and development and it also contributes to plant defense. Auxin activates downstream signaling by promoting degradation of the repressor Aux/IAA proteins to liberate the key transcription factors ARFs (auxin response factors). Pathogens have been known to mitigate plant defense by targeting Aux/IAA proteins to manipulate plant auxin signaling , but whether ARFs are also targeted by pathogens was unknown. Zhang et al. found that plant viruses target OsARF17, a rice ARF, in multiple ways. They showed that proteins of various types of viruses, including ssRNA viruses, a dsRNA virus, and a negative-sense ssRNA virus interact with different domains of OsARF17, and interfere with its transcriptional or DNA binding activity. Analyses using OsARF17 overexpressing or knock-out plants showed that OsARF17 is a positive regulator of plant defense against these distinct types of plant viruses. This study reveals that OsARF17 plays an important role in defense against various plant viruses, and suggests that distinct viral pathogens have independently evolved strategies to target this key component of auxin signaling to facilitate infection. (Summary by Tatsuya Nobori) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1918254117
Auxin regulates various aspects of plant growth and development and it also contributes to plant defense. Auxin activates downstream signaling by promoting degradation of the repressor Aux/IAA proteins to liberate the key transcription factors ARFs (auxin response factors). Pathogens have been known to mitigate plant defense by targeting Aux/IAA proteins to manipulate plant auxin signaling , but whether ARFs are also targeted by pathogens was unknown. Zhang et al. found that plant viruses target OsARF17, a rice ARF, in multiple ways. They showed that proteins of various types of viruses, including ssRNA viruses, a dsRNA virus, and a negative-sense ssRNA virus interact with different domains of OsARF17, and interfere with its transcriptional or DNA binding activity. Analyses using OsARF17 overexpressing or knock-out plants showed that OsARF17 is a positive regulator of plant defense against these distinct types of plant viruses. This study reveals that OsARF17 plays an important role in defense against various plant viruses, and suggests that distinct viral pathogens have independently evolved strategies to target this key component of auxin signaling to facilitate infection. (Summary by Tatsuya Nobori) Proc. Natl. Acad. Sci. USA 10.1073/pnas.1918254117
Ploidy affects the seed, dormancy and seedling characteristics of a perennial grass, conferring an advantage in stressful climates ($)
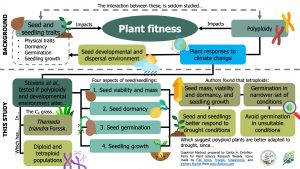 Both polyploidy (i.e., having more than two sets of chromosomes) and seed traits influence plant fitness. However, the interaction between these two factors is underexplored. Here, Stevens et al. tested the effect of polyploidy and seed developmental environment on 1) seed viability and mass; 2) seed dormancy; 3) seed germination under drought; and 4) seedling growth. To achieve this, they used Themeda triandra –an Australian C4 grass– seeds from diploid and tetraploid accessions, which came from three different regions and were exposed to either ambient or experimental warming conditions. Despite the fact that the region and the warming treatment differentially affected seed traits, the authors found tetraploid seeds had higher mass, viability, and dormancy in general. Also, seedlings from tetraploid plants were larger and grew faster. However, tetraploid seeds germinated on a narrower set of conditions. These results indicate that polyploid plants are better adapted to drought by having seedlings that perform better under drought and restricting germination in unsuitable conditions –via seed dormancy and stricter germination requirements. (Summary by Carlos A. Ordóñez-Parra) Plant Biol. 10.1111/plb.13094
Both polyploidy (i.e., having more than two sets of chromosomes) and seed traits influence plant fitness. However, the interaction between these two factors is underexplored. Here, Stevens et al. tested the effect of polyploidy and seed developmental environment on 1) seed viability and mass; 2) seed dormancy; 3) seed germination under drought; and 4) seedling growth. To achieve this, they used Themeda triandra –an Australian C4 grass– seeds from diploid and tetraploid accessions, which came from three different regions and were exposed to either ambient or experimental warming conditions. Despite the fact that the region and the warming treatment differentially affected seed traits, the authors found tetraploid seeds had higher mass, viability, and dormancy in general. Also, seedlings from tetraploid plants were larger and grew faster. However, tetraploid seeds germinated on a narrower set of conditions. These results indicate that polyploid plants are better adapted to drought by having seedlings that perform better under drought and restricting germination in unsuitable conditions –via seed dormancy and stricter germination requirements. (Summary by Carlos A. Ordóñez-Parra) Plant Biol. 10.1111/plb.13094
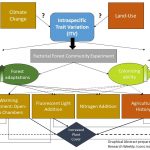
Individualistic responses of forest herb traits to environmental change ($)
 Functional traits have shown promise for their ability to improve predictions of interspecific plant responses to environmental factors, but most trait-based studies have not addressed the role of intraspecific trait variation (ITV) in trait-environment relationships. ITV may be directly or indirectly altered by climate change pressures and land use histories, depending on individual levels of phenotypic plasticity. Blondeel et al. use a factorial manipulative experiment to test changes in mean plant height and mean specific leaf area (SLA) within 15 different plant species from temperate European forests in response to warming with open-top chambers, fluorescent light addition, and nitrogen addition. Community members were diversified to include a range of levels of forest (shade) adaptation and colonization ability and soils had different agricultural use histories. ITV responses to treatments were variable and dependent on species but some patterns did emerge. This study found that mean plant height increased within species in response to more available light, warming, and agricultural land-use due largely to enhanced vegetative cover. The mean plant height of fast-colonizers increased the most in response to treatments. These results highlight the potential importance of the indirect effects of climate change and land use history in the form of increased competition and a shift towards fast growing and colonizing species. Surprisingly, there were no interactive effects of treatments on ITV and mean SLA experienced species-dependent bidirectional change with few strong patterns emerging in response to treatments. (Summary by Sienna Wessel) Plant Biology 10.1111/plb.13103
Functional traits have shown promise for their ability to improve predictions of interspecific plant responses to environmental factors, but most trait-based studies have not addressed the role of intraspecific trait variation (ITV) in trait-environment relationships. ITV may be directly or indirectly altered by climate change pressures and land use histories, depending on individual levels of phenotypic plasticity. Blondeel et al. use a factorial manipulative experiment to test changes in mean plant height and mean specific leaf area (SLA) within 15 different plant species from temperate European forests in response to warming with open-top chambers, fluorescent light addition, and nitrogen addition. Community members were diversified to include a range of levels of forest (shade) adaptation and colonization ability and soils had different agricultural use histories. ITV responses to treatments were variable and dependent on species but some patterns did emerge. This study found that mean plant height increased within species in response to more available light, warming, and agricultural land-use due largely to enhanced vegetative cover. The mean plant height of fast-colonizers increased the most in response to treatments. These results highlight the potential importance of the indirect effects of climate change and land use history in the form of increased competition and a shift towards fast growing and colonizing species. Surprisingly, there were no interactive effects of treatments on ITV and mean SLA experienced species-dependent bidirectional change with few strong patterns emerging in response to treatments. (Summary by Sienna Wessel) Plant Biology 10.1111/plb.13103