Plant 22-nt siRNAs mediate translational repression and stress adaptation (Nature)
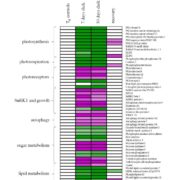
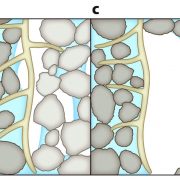
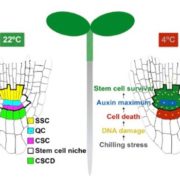
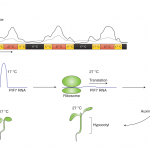
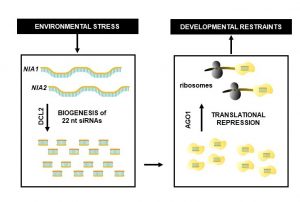 Among the myriad small interfering RNAs, 21- and 24-nucleotides siRNAs control plant development and immunity through mRNA cleavage and RNA-directed DNA methylation, respectively. Still, the regulation and biological function of 22-nt siRNAs remain unresolved. In this report, Wu and coworkers investigated the massive accumulation of 22-nt siRNAs in plants with mutations in DICER-like 4 (DCL4) and RNA-degrading enzymes ETHYLENE INSENSITIVE5 (EIN5) and SUPER KILLER2 (SKI2), and found that two genes encoding nitrate reductases (NIA1, NIA2) contributed to almost half of the total amount of 22-nt siRNAs. Noteworthy, transcriptome analysis and polysome profiling of NIA1/2 revealed that 22-nt siRNAs repress translation globally as well as gene-specifically. Plants that are blocked in RNA decay and accumulate 22-nt siRNAs (ein5 dcl4 and ski2 dcl4) display root growth inhibition, abnormal phloem development and anthocyanin accumulation. Similar pleiotropic defects and accumulation of 22-nt siRNAs were also observed in wild-type plants in response to abiotic stress (i.e., nitrogen starvation, high salinity and ABA treatment), indicating a key role for 22-nt siRNAs in the fine-tuning of stress adaptation. To conclude, stress leading to an increase in 22-nt siRNAs likely causes a shortage of amino acids mediated by translational repression of NIA1/2, and consequently the slowdown of global translation leading to developmental restraint. Thus, this strategy allows plants to switch from growth to defense under environmental limitations. (Summary by Michela Osnato @michela_osnato) Nature 10.1038/s41586-020-2231-y
Among the myriad small interfering RNAs, 21- and 24-nucleotides siRNAs control plant development and immunity through mRNA cleavage and RNA-directed DNA methylation, respectively. Still, the regulation and biological function of 22-nt siRNAs remain unresolved. In this report, Wu and coworkers investigated the massive accumulation of 22-nt siRNAs in plants with mutations in DICER-like 4 (DCL4) and RNA-degrading enzymes ETHYLENE INSENSITIVE5 (EIN5) and SUPER KILLER2 (SKI2), and found that two genes encoding nitrate reductases (NIA1, NIA2) contributed to almost half of the total amount of 22-nt siRNAs. Noteworthy, transcriptome analysis and polysome profiling of NIA1/2 revealed that 22-nt siRNAs repress translation globally as well as gene-specifically. Plants that are blocked in RNA decay and accumulate 22-nt siRNAs (ein5 dcl4 and ski2 dcl4) display root growth inhibition, abnormal phloem development and anthocyanin accumulation. Similar pleiotropic defects and accumulation of 22-nt siRNAs were also observed in wild-type plants in response to abiotic stress (i.e., nitrogen starvation, high salinity and ABA treatment), indicating a key role for 22-nt siRNAs in the fine-tuning of stress adaptation. To conclude, stress leading to an increase in 22-nt siRNAs likely causes a shortage of amino acids mediated by translational repression of NIA1/2, and consequently the slowdown of global translation leading to developmental restraint. Thus, this strategy allows plants to switch from growth to defense under environmental limitations. (Summary by Michela Osnato @michela_osnato) Nature 10.1038/s41586-020-2231-y
[altmetric doi=”10.1038/s41586-020-2231-y” details=”right” float=”right”]