Phloem Companion Cell-Specific Transcriptomics and Epigenomics
By conducting phloem companion cell-specific transcriptomic and epigenomic analyses, You et al. identify MRF1, a novel regulator of flowering. Plant Cell https://doi.org/10.1105/tpc.17.00714
By Yuan You1,2 and Markus Schmid1,3
1 Max Planck Institute for Developmental Biology, Department of Molecular Biology, Max-Planck-Ring 9, 72076 Tübingen, Germany.
2 Center for Plant Molecular Biology (ZMBP), Department of General Genetics, University Tübingen, Auf der Morgenstelle 32, 72076 Tübingen, Germany.
3 Umeå Plant Science Centre, Department of Plant Physiology, Umeå University, SE-901 87 Umeå, Sweden.
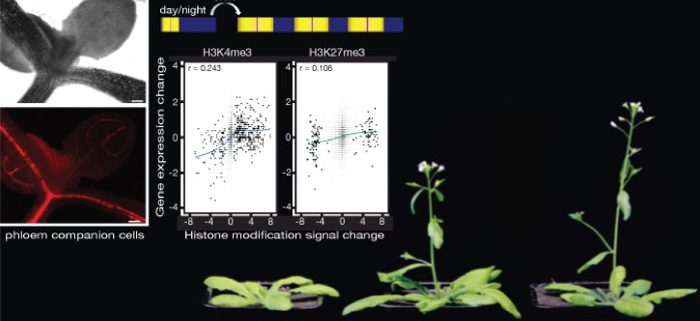
Background: In vascular plants, the phloem distributes soluble sugars and signalling molecules throughout the plant. The phloem consists of three cell types: phloem supportive cells, sieve elements (SE), and phloem companion cells (PCCs). The latter uploads soluble sugars synthesised in the leaf mesophyll and signal molecules into SEs, by which they are transported to other parts of the plant. Thus, PCCs play a central role in coordinating growth and development processes in response to environmental changes.
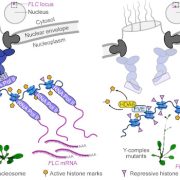
Question: How do chromatin modifications, such as tri-methylations at lysine 4 and lysine 27 of histone protein H3 (H3K4me3 and H3K27me3), contribute to the regulation of gene expression in Arabidopsis thaliana PCCs in response to daylength changes?
Findings: We performed a genome-wide analysis of epigenome and transcriptome responses in Arabidopsis PCCs following a change in daylength. Genes that responded to the inductive long day, which promotes flowering in Arabidopsis, included key components of the circadian clock and regulators of development, metabolism, and chromatin modifications. H3K4me3 at the transcription start site (TSS) is positively associated with active gene expression in PCCs. By contrast, associations between H3K27me3 and gene expression are much more complex. In combination with H3K4me3 at the TSS, changes in H3K27me3 signals are positively correlated with changes in gene expression. This is unexpected, as this mark is usually associated with repressed genes. By itself, H3K27me3 appears to contribute little to gene expression regulation. The dynamic changes of these two important histone modifications illustrated the complex gene-regulatory networks that integrate plant developmental and physiological processes in PCCs. Finally, our cell type-specific analyses identified MRF1, a previously uncharacterized MORN-motif repeat protein, as both necessary and sufficient to promote flowering. Molecular analyses suggest that MRF1 acts upstream of FT, a central integrator of the photoperiod pathway.
Next steps: Our findings emphasize the need for cell type-specific transcriptome and epigenome studies to clarify the complex molecular regulatory networks of gene expression and chromatin landscape that govern a plant’s responses to environmental changes. It will be interesting to examine the molecular functions of MRF1 in the canonical flowering time pathways in more detail.
Yuan You, Aneta Sawikowska, Joanne E. Lee, Ruben M. Benstein, Manuela Neumann, Paweł Krajewski, and Markus Schmid. (2019). Phloem Companion Cell-specific Transcriptomic and Epigenomic Analyses Identify MRF1, a Regulator of Flowering. https://doi.org/10.1105/tpc.17.00714
Key words: Arabidopsis phloem companion cells, transcriptomics and epigenomics, INTACT, MORN-motif repeat protein