Personal Trainer: bHLH121 Functions Upstream of a Transcriptional Network of Heavy Lifters Involved in Balancing Iron Levels
Iron is a cofactor of numerous plant proteins that function in processes ranging from redox reactions during photosynthesis to respiration. Typical soils contain approximately 1–5% iron, but much of this iron is not readily accessible to plants due to low solubility. Nongraminaceous plants help solubilize iron (Fe3+) in the soil by making the rhizosphere more acidic and secreting Fe3+-mobilizing coumarins. The plant takes up iron from this solubilized Fe3+ pool after generating reduced Fe2+ at the root surface via the activity of enzymes such as FERRIC REDUCTION OXIDASE2 (FRO2). The reduced Fe2+ ions are transported across the plasma membrane into the root by IRON-REGULATED TRANSPORTER1 (IRT1). Once inside the plant, the iron is transported to the proper locations, stored, and used as needed (reviewed in Kobayashi and Nishizawa, 2012).
The properties that make iron so suitable for redox reactions also make it readily generate harmful reactive oxygen species. Therefore, plants must maintain the appropriate iron levels by sensing iron status throughout the plant and responding accordingly. Iron sensors, which have yet to be identified, activate long-distance signaling from the shoot (or short-distance signaling throughout the root) through various pathways, triggering transcriptional reprogramming to alter iron status. Thus, sensors (or closely associated proteins) must be present throughout the plant regardless of iron status and orchestrate the movement of iron to the appropriate locations as needed.
 In Arabidopsis thaliana (Arabidopsis), changes in iron status trigger the activity of an intricate network of at least 16 bHLH transcription factors (TFs) that regulate iron homeostasis (Gao et al., 2019). Two of these TFs, FIT (FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR) and ILR3 (IAA-LEUCINE RESISTANT3), function as crucial nodes in the Fe homeostasis network by regulating the expression of heavy lifters such as IRT1 and FRO2. How these network branches act in concert with each other and with upstream factors such as iron sensors has remained elusive, but Gao et al. (2020) made great strides in answering these questions.
In Arabidopsis thaliana (Arabidopsis), changes in iron status trigger the activity of an intricate network of at least 16 bHLH transcription factors (TFs) that regulate iron homeostasis (Gao et al., 2019). Two of these TFs, FIT (FER-LIKE IRON DEFICIENCY-INDUCED TRANSCRIPTION FACTOR) and ILR3 (IAA-LEUCINE RESISTANT3), function as crucial nodes in the Fe homeostasis network by regulating the expression of heavy lifters such as IRT1 and FRO2. How these network branches act in concert with each other and with upstream factors such as iron sensors has remained elusive, but Gao et al. (2020) made great strides in answering these questions.
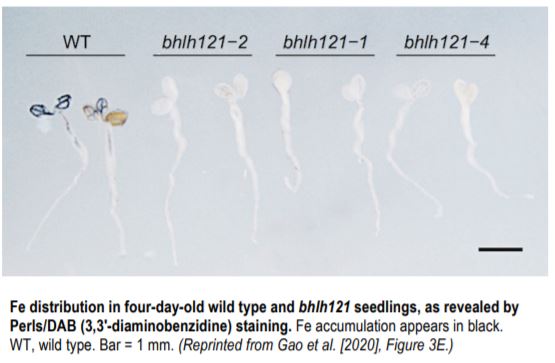
The authors performed co-immunoprecipitation analysis followed by mass spectrometry to search for interacting partners of ILR3 and identified bHLH121, a close homolog of POPEYE. The interaction of bHLH121 with ILR3 and its close homologs was confirmed in various assays. bhlh121 loss-of-function mutants showed reduced tolerance to low iron levels and abnormally low iron contents in seeds and young seedlings (see Figure). The transcript levels of iron uptake genes FRO2 and IRT1, as well as genes involved in coumarin biosynthesis and secretion, were reduced in the mutants. Expression analysis of bhlh121 and wild-type plants in the presence and absence of iron deficiency helped position bHLH121 high up in the transcriptional regulatory cascade of iron deficiency responses, as confirmed by analysis of transgenic plants overexpressing bHLH121 driven by the UBIQUITIN10 promoter.
To explore the localization of bHLH121, the authors generated transgenic bhlh121 plants whose loss-of-function phenotypes were rescued by the promoter-containing genomic sequence of bHLH121 fused to the GFP reporter gene and tracked the expression of the bHLH121-GFP fusion protein in cell nuclei. The fusion protein was expressed throughout the plant body, a localization not affected by Fe availability. By contrast, Fe availability affected the types of cells in roots containing bHLH12. Under Fe sufficiency, bHLH121 was mainly detected in the nuclei of cells in the stele, whereas under Fe deficiency, most bHLH121 was observed in the root cortex and rhizodermis cells. Therefore, perhaps bHLH121 is a long-sought regulator of plant responses to iron deficiency that functions upstream of FIT in concert with ILR3 and its closest homologs, placing it within striking distance of iron sensors.
Jennifer Lockhart
Science Editor
ORCID: 0000-0002-1394-8947
REFERENCES
Gao, F., Robe, K., Bettembourg, M., Navarro, N., Rofidal, V., Santoni, V., Gaymard, F., Vignols, F., Roschzttardtz, H., Izquierdo, E., and Dubos, C. (2020).
The transcription factor bHLH121 interacts with bHLH105 (ILR3) and its closest homologs to regulate iron homeostasis in Arabidopsis. Plant Cell 32: doi:10.1015/tpc19.00541. {https://doi.org/10.1105/tpc.19.00541}
Gao, F., Robe, K., Gaymard, F., Izquierdo, E., and Dubos, C. (2019). The transcriptional control of iron homeostasis in plants: a tale of bHLH transcription factors?. Front Plant Sci 10: 6.
Kobayashi, T. and Nishizawa, N.K. (2012). Iron uptake, translocation, and regulation in higher plants. Ann Rev Plant Biol 63: 131-152.