How To Identify Autophagy Modulators
Author: Masanori Izumi
Affiliation: RIKEN Center for Sustainable Resource Science, Wako 351-0198, Japan
Autophagy is the ubiquitous process in eukaryotes leading to the degradation of intracellular components, during which a portion of the cytoplasm, including organelles, is sequestered by nascent autophagic membranes and delivered into the lytic organelles for degradation (Mizushima and Komatsu, 2011). Over the past decade, the roles of autophagy in plants have been extensively studied. Numerous reports demonstrate an essential role of plant autophagy in facilitating nutrient recycling during developmental growth or adaptation to starvation (Marshall and Vierstra, 2018). Plant autophagy can remove selective targets such as denatured proteins or damaged organelles and thereby maintain cellular homeostasis. The accumulating evidence reveals that autophagy is a fundamental process supporting plant growth and crop productivity (Marshall and Vierstra, 2018).
Forward genetics approaches using the budding yeast Saccharomyces cerevisiae originally identified autophagy-related (ATG) genes (Takeshige et al., 1992; Tsukada and Ohsumi, 1993). Mutant analysis of plant orthologues of the yeast ATGs advanced our understanding of plant autophagy (Yoshimoto and Ohsumi, 2018). However, such genetic approaches have limitations; for instance, it is challenging to identify redundant genes or embryo-essential genes. Recently, approaches involving screening plant responses to libraries of small molecules have been developed to identify the molecular basis for various processes (Dejonghe and Russinova, 2017; Kinoshita et al., 2018). In the current issue of Plant Physiology, Dauphinee et al. (2019) established a high-throughput screening strategy to identify novel autophagy modulators in plants.
AUTOPHAGY8 (ATG8) is a key protein of autophagy that is incorporated into autophagic membranes and is degraded along with the targets (Ichimura et al., 2000). The current screening method comprises four steps using luciferase (Luc)- or fluorescent protein-labeled ATG8 proteins: i) detection of Luc-ATG8-derived signals in tobacco (Nicotiana tabacum) BY2 cells; ii) ratio imaging of tandem fluorescent proteins fused to ATG8; iii) immunoblot detection of green fluorescent protein (GFP) released from the cleavage of GFP-ATG8; and iv) assessment of off-target effects (Fig. 1). To evaluate the validity of the method, the authors tested some small chemical libraries in the screening pipeline, which identified a candidate plant autophagy activator.
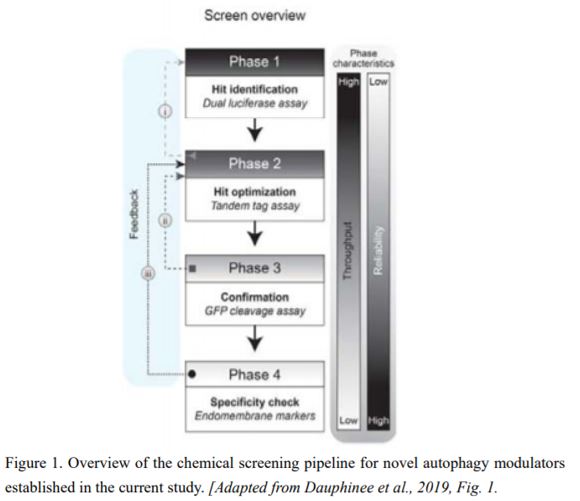 In the current method, Dauphinee et al. (2019) used two types of ratio imaging, Firefly Luc to Renilla reniformis Luc and red fluorescent protein (RFP) to GFP, at the early stage of the pipeline, where large numbers of chemicals are assessed. At the first stage, the signal from Firefly Luc fused to ATG8 is normalized to nucleus-targeted Renilla Luc signal to evaluate the reduction of Luc-ATG8 signals caused by their vacuolar digestion. The second stage takes place in transgenic Arabidopsis thaliana plants expressing ATG8 proteins tagged by dual fluorescent reporters, RFP and GFP. The ratio of RFP to GFP is measured via confocal microscopy, and since RFP is relatively stable compared to GFP in vacuolar lytic activity, an increase in the RFP/GFP ratio represents activation of autophagy (Kimura et al., 2007). Such ratio imaging helps eliminate bias due to factors such as differences in cell volume, allowing for efficient, large-scale chemical screening. Step iii is a widely used method to evaluate autophagy flux (Mizushima et al., 2010), which can robustly show the effect of the screened candidates. In the final step, the authors attempted to remove chemicals that affect autophagy activity through off-target effects.
In the current method, Dauphinee et al. (2019) used two types of ratio imaging, Firefly Luc to Renilla reniformis Luc and red fluorescent protein (RFP) to GFP, at the early stage of the pipeline, where large numbers of chemicals are assessed. At the first stage, the signal from Firefly Luc fused to ATG8 is normalized to nucleus-targeted Renilla Luc signal to evaluate the reduction of Luc-ATG8 signals caused by their vacuolar digestion. The second stage takes place in transgenic Arabidopsis thaliana plants expressing ATG8 proteins tagged by dual fluorescent reporters, RFP and GFP. The ratio of RFP to GFP is measured via confocal microscopy, and since RFP is relatively stable compared to GFP in vacuolar lytic activity, an increase in the RFP/GFP ratio represents activation of autophagy (Kimura et al., 2007). Such ratio imaging helps eliminate bias due to factors such as differences in cell volume, allowing for efficient, large-scale chemical screening. Step iii is a widely used method to evaluate autophagy flux (Mizushima et al., 2010), which can robustly show the effect of the screened candidates. In the final step, the authors attempted to remove chemicals that affect autophagy activity through off-target effects.
Once chemicals that modulate autophagy have been identified, researchers need to identify their target molecules. Although such target identification can be challenging, doing so may allow us to understand novel mechanisms regulating plant autophagy. Furthermore, as the importance of autophagy in the productivity of crops such as rice (Oryza sativa) and maize (Zea mays) is well established, identifying autophagy-specific modulators might suggest a strategy to develop new agricultural chemicals that enhance productivity or stress tolerance through controlling autophagy activity (Wada et al., 2015; McLoughlin et al., 2018).
LITERATURE CITED
Dauphinee AN, Cardoso C, Dalman K, Ohlsson JA, Berglund Fick S, Robert S, Hicks GR, Bozhkov P, Minina EA (2019) Chemical screening pipeline for identification of specific plant autophagy modulators. Plant Physiol https://doi.org/10.1104/pp.19.00647
Dejonghe W, Russinova E (2017) Plant Chemical Genetics: From Phenotype-Based Screens to Synthetic Biology. Plant Physiol 174: 5-20
Ichimura Y, Kirisako T, Takao T, Satomi Y, Shimonishi Y, Ishihara N, Mizushima N, Tanida I, Kominami E, Ohsumi M, Noda T, Ohsumi Y (2000) A ubiquitin-like system mediates protein lipidation. Nature 408: 488-492
Kimura S, Noda T, Yoshimori T (2007) Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy 3: 452-460
Kinoshita T, McCourt P, Asami T, Torii KU (2018) Plant Chemical Biology. Plant Cell Physiol 59: 1483-1486
Marshall RS, Vierstra RD (2018) Autophagy: The master of bulk and selective recycling. Annu Rev Plant Biol 69: 173-208
McLoughlin F, Augustine RC, Marshall RS, Li FQ, Kirkpatrick LD, Otegui MS, Vierstra RD (2018) Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants 4: 1056-1070
Mizushima N, Komatsu M (2011) Autophagy: renovation of cells and tissues. Cell 147: 728-741
Mizushima N, Yoshimori T, Levine B (2010) Methods in mammalian autophagy research. Cell 140: 313-326
Takeshige K, Baba M, Tsuboi S, Noda T, Ohsumi Y (1992) Autophagy in yeast demonstrated with proteinase-deficient mutants and conditions for its induction. J Cell Biol 119: 301-311
Tsukada M, Ohsumi Y (1993) Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett 333: 169-174
Wada S, Hayashida Y, Izumi M, Kurusu T, Hanamata S, Kanno K, Kojima S, Yamaya T, Kuchitsu K, Makino A, Ishida H (2015) Autophagy supports biomass production and nitrogen use efficiency at the vegetative stage in rice. Plant Physiol 168: 60-73
Yoshimoto K, Ohsumi Y (2018) Unveiling the molecular mechanisms of plant autophagy-from autophagosomes to vacuoles in plants. Plant Cell Physiol 59: 1337-1344