Pan-genome analysis of 33 genetically diverse rice accessions reveals hidden genomic variations (Cell)
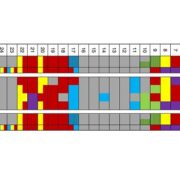
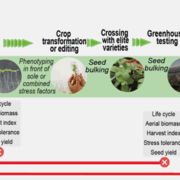
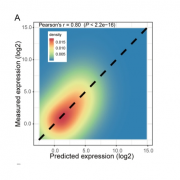
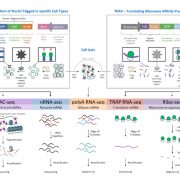
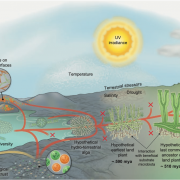
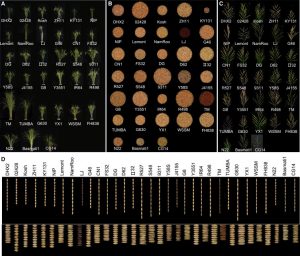 In spite of the considerable excitement that accompanies the first genomic sequence of a species, that sequence usually represents a single individual or inbred accession – the real excitement comes from comparisons between individuals or accessions. These pan-genomic analyses provide insights into the sources of genetic variability. Here, Qin et al. compare the genomes of 33 diverse rice accessions: 32 Orzya sativa and 1 Oryza glaberrima, African cultivated rice, which was domesticated independently and serves as an outgroup to help to identify ancestral versus derived variations. This comparative analysis demonstrated numerous Structural Variations (SVs) including Presence/Absence Variations (PAVs), as well as gene Copy Number Variations (gCNVs). Both SVs and gCNVs are correlated with variations in gene expression levels and agronomic traits. The authors produced a graph-based genome that includes the full genetic diversity from these diverse accessions, providing a rich and powerful dataset that can be browsed here http://ricerc.sicau.edu.cn/. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2021.04.046
In spite of the considerable excitement that accompanies the first genomic sequence of a species, that sequence usually represents a single individual or inbred accession – the real excitement comes from comparisons between individuals or accessions. These pan-genomic analyses provide insights into the sources of genetic variability. Here, Qin et al. compare the genomes of 33 diverse rice accessions: 32 Orzya sativa and 1 Oryza glaberrima, African cultivated rice, which was domesticated independently and serves as an outgroup to help to identify ancestral versus derived variations. This comparative analysis demonstrated numerous Structural Variations (SVs) including Presence/Absence Variations (PAVs), as well as gene Copy Number Variations (gCNVs). Both SVs and gCNVs are correlated with variations in gene expression levels and agronomic traits. The authors produced a graph-based genome that includes the full genetic diversity from these diverse accessions, providing a rich and powerful dataset that can be browsed here http://ricerc.sicau.edu.cn/. (Summary by Mary Williams @PlantTeaching) Cell 10.1016/j.cell.2021.04.046