Rafael Massahiro Yassue: Plant Direct First Author

 Rafael Massahiro Yassue, first author of “Genome-wide association analysis of hyperspectral reflectance data to dissect the genetic architecture of growth-related traits in maize under plant growth-promoting bacteria inoculation”
Rafael Massahiro Yassue, first author of “Genome-wide association analysis of hyperspectral reflectance data to dissect the genetic architecture of growth-related traits in maize under plant growth-promoting bacteria inoculation”
Current Position: Data Scientist at GDM
Education: Ph.D. in Genetics and Plant Breeding, University of São Paulo (ESALQ/USP), Brazil
Non-scientific Interests: Hiking, traveling, and cooking
Brief bio: As an Agronomist with a Ph.D. in genetics and plant breeding, my thesis focused on utilizing cutting-edge technologies to connect genomics, phenomics, and statistical modeling in plant breeding and genetics studies. The study involved using high-throughput phenotyping techniques, such as hyperspectral, thermal, and multispectral cameras, as well as RGB imaging, to understand the genetic architecture of the response to plant growth-promoting bacteria (PGPB) in tropical maize. The research explores the use of these advanced phenotyping methods to identify the root and shoot traits and their genetic basis, and how they were associated with the response to PGPB. Currently, I am a Data Scientist at GDM. My research interest is using experimental design, genomic selection, phenomics, and enviromics to enhance genetic gain in the context of Soybean breeding.



 Chaofan Chen, first author of
Chaofan Chen, first author of  Takatoshi Kiba, first author of
Takatoshi Kiba, first author of 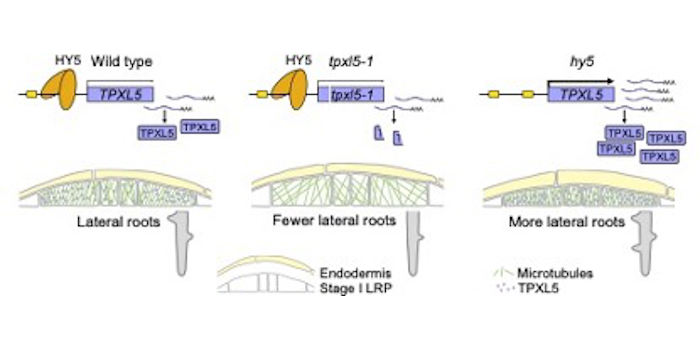 Findings: We found that a microtubule-stabilizing protein TPX2-LIKE5 (TPXL5) participates in LR initiation. In the tpxl5 mutant, the radial expansion in the peripheral domain of LR founder cells after the first asymmetric division was stronger than that in wildtype, and the ordered transverse cortical microtubule arrays were not well generated. Moreover, transcription factor ELONGATED HYPOCOTYL5 (HY5) downregulated TPXL5 expression. Mutant hy5 exhibited the opposite phenotype compared to the tpxl5 mutant. Our study demonstrates that TPXL5 positively regulates cortical microtubules reorganization in the peripheral domain, thus promoting the asymmetric radial expansion of founder cells during LR initiation. It also reveals a novel molecular mechanism by which HY5 regulates TPXL5-mediated cortical microtubule reorganization and cell remodeling during LR initiation.
Findings: We found that a microtubule-stabilizing protein TPX2-LIKE5 (TPXL5) participates in LR initiation. In the tpxl5 mutant, the radial expansion in the peripheral domain of LR founder cells after the first asymmetric division was stronger than that in wildtype, and the ordered transverse cortical microtubule arrays were not well generated. Moreover, transcription factor ELONGATED HYPOCOTYL5 (HY5) downregulated TPXL5 expression. Mutant hy5 exhibited the opposite phenotype compared to the tpxl5 mutant. Our study demonstrates that TPXL5 positively regulates cortical microtubules reorganization in the peripheral domain, thus promoting the asymmetric radial expansion of founder cells during LR initiation. It also reveals a novel molecular mechanism by which HY5 regulates TPXL5-mediated cortical microtubule reorganization and cell remodeling during LR initiation.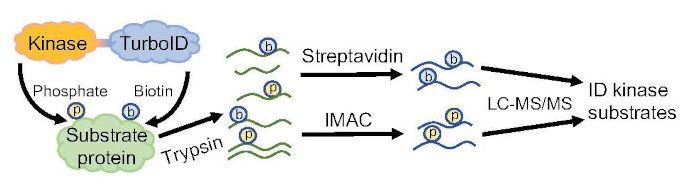 Findings: We show that TurboID is an effective and specific tool for mapping kinase signaling networks. We identified 482 BIN2 proximal proteins, including about two-thirds that showed BIN2-dependent phosphorylation and many known BIN2 interactors and substrates. The dataset of in vivo BIN2 interactors and substrates uncovers an expansive signaling network and reveals a convergence between the BIN2/GSK3 and O-GlcNAc modification pathways in both plants and animals.
Findings: We show that TurboID is an effective and specific tool for mapping kinase signaling networks. We identified 482 BIN2 proximal proteins, including about two-thirds that showed BIN2-dependent phosphorylation and many known BIN2 interactors and substrates. The dataset of in vivo BIN2 interactors and substrates uncovers an expansive signaling network and reveals a convergence between the BIN2/GSK3 and O-GlcNAc modification pathways in both plants and animals.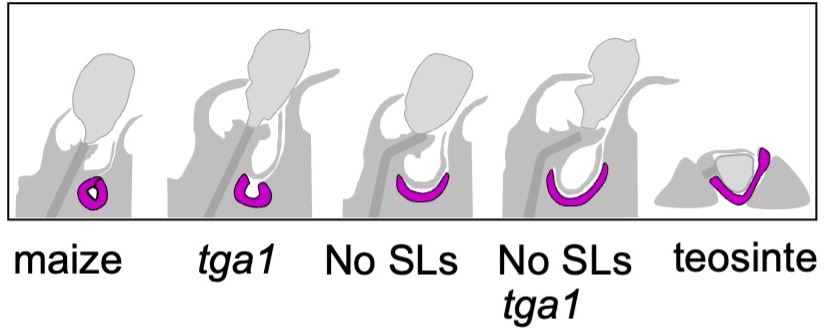 Findings: A rare combination of domestication features emerged in maize mutants unable to form or sense SLs. Kernels were small, cobs primitive, and seed-bearing cupules larger. Also, when SL-deficient mutants were combined with an ancestral form of the known domestication gene, Tga1 (Teosinte glume architecture 1), ears and kernels showed the most primitive, teosinte-like features yet observed. In addition, without SLs coordinating development, rigid seed-bearing cupules often ruptured growing kernels. Finally, analysis of underlying mechanisms indicated that SLs could act alone as well as regulate Tga1 by sequestering its protein. Evidence included RNA-seq expression analysis, protein-protein interactions, and additional maize genetics.
Findings: A rare combination of domestication features emerged in maize mutants unable to form or sense SLs. Kernels were small, cobs primitive, and seed-bearing cupules larger. Also, when SL-deficient mutants were combined with an ancestral form of the known domestication gene, Tga1 (Teosinte glume architecture 1), ears and kernels showed the most primitive, teosinte-like features yet observed. In addition, without SLs coordinating development, rigid seed-bearing cupules often ruptured growing kernels. Finally, analysis of underlying mechanisms indicated that SLs could act alone as well as regulate Tga1 by sequestering its protein. Evidence included RNA-seq expression analysis, protein-protein interactions, and additional maize genetics.
 Shugang Hui, first author of
Shugang Hui, first author of 
 Ana Lopez Vazquez, first author of
Ana Lopez Vazquez, first author of