Mapping the BIN2 kinase signaling network with TurboID-mediated proximity labeling and phosphoproteomics
Background: Intracellular signal transduction relies on specific and dynamic interactions between kinases and their substrates. Identifying substrate proteins of each kinase is crucial for understanding cellular signaling transduction pathways but is technically challenging because of the transient nature of the enzyme-substrate interactions and the large number of kinases acting in a cell. Arabidopsis thaliana BRASSINOSTEROID-INSENSITIVE2 (BIN2) is one of the best-studied plant kinases, with key roles in multiple signaling pathways including the brassinosteroid and auxin pathways. However, BIN2’s in vivo interactors and substrate proteins have not been fully characterized. Recent studies have developed the TurboID biotin ligase as a highly efficient proximity labeling tool; its efficiency in mapping transient protein-protein interactions has not been fully explored.
Question: Can a fusion protein containing a kinase and the TurboID biotin ligase biotinylate the substrate proteins phosphorylated by the BIN2 kinase? Is this approach effective, when combined with phosphoproteomics, in identifying kinase substrates that interact transiently? What are the substrates and cellular targets of BIN2? How does the BIN2 signaling network overlap with other signaling pathways?
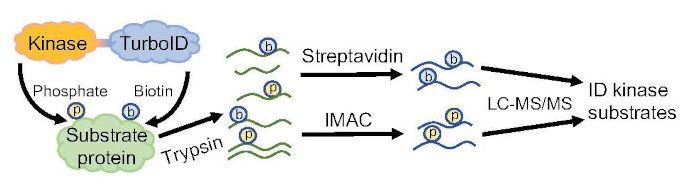 Findings: We show that TurboID is an effective and specific tool for mapping kinase signaling networks. We identified 482 BIN2 proximal proteins, including about two-thirds that showed BIN2-dependent phosphorylation and many known BIN2 interactors and substrates. The dataset of in vivo BIN2 interactors and substrates uncovers an expansive signaling network and reveals a convergence between the BIN2/GSK3 and O-GlcNAc modification pathways in both plants and animals.
Findings: We show that TurboID is an effective and specific tool for mapping kinase signaling networks. We identified 482 BIN2 proximal proteins, including about two-thirds that showed BIN2-dependent phosphorylation and many known BIN2 interactors and substrates. The dataset of in vivo BIN2 interactors and substrates uncovers an expansive signaling network and reveals a convergence between the BIN2/GSK3 and O-GlcNAc modification pathways in both plants and animals.
Next steps: How BIN2 acts specifically in various signaling pathways and how it regulates various substrate proteins and cellular functions are key questions to be answered in future studies. How BIN2-mediated phosphorylation crosstalks with O-GlcNAcylation is another important question with broad implications. Our dataset of candidate proteins with modification sites will enable future investigations that advance our understanding of these important questions.
Reference:
Tae-Wuk Kim, Chan Ho Park, Chuan-Chih Hsu, Yeong-Woo Kim, Yeong-Woo Ko, Zhenzhen Zhang, Jia-Ying Zhu, Yu-Chun Hsiao, Tess Branon, Krista Kaasik, Evan Saldivar, Kevin Li, Asher Pasha, Nicholas J Provart, Alma L Burlingame, Shou-Ling Xu, Alice Y Ting, Zhi-Yong Wang (2023) Mapping the signaling network of BIN2 kinase using TurboID-mediated biotin labeling and phosphoproteomics. https://doi.org/10.1093/plcell/koad013