IN BRIEF by Jennifer Lockhart [email protected]
Plant cell components that are no longer needed are degraded in the vacuole, but they don’t get there by magic. Sack-like double-membrane structures called autophagosomes engulf this cellular rubbish and neatly transport it to the vacuole for degradation. Autophagy, a highly conserved process orchestrated by a suite of evolutionarily conserved autophagy-related proteins (ATGs), goes into high gear under stress conditions to quickly recycle nutrients and maintain cellular homeostasis (reviewed in Liu and Bassham, 2012). In mammals, upon autophagy induction, the scaffold protein Beclin-1 becomes ubiquitinated by E3 ligases such as Tumor necrosis factor Receptor-Associated Factor (TRAF)-family proteins. Ubiquitinated Beclin-1 stimulates the kinase activity of VPS34, thereby generating the phospholipid PI3P, which acts as a docking site for other components, leading to autophagosome formation. In addition to serving as E3 ligases, TRAF-family proteins play key roles as adaptors in diverse signaling cascades (reviewed in Wajant et al., 2001).
Despite the crucial role of ubiquitination in mammalian autophagosome formation, little is known about this process in plants. Prompted by the notion that TRAF-family proteins might be critical for autophagy in plants as well as mammals, Qi et al. (2017) investigated this family in Arabidopsis thaliana and uncovered an intriguing mechanism for the regulation of plant autophagosome formation. Two proteins, TRAF1a and TRAF1b, localized to the cytoplasm in protoplasts transiently expressing tagged TRAF fusion proteins under normal conditions but to autophagosomes under starvation conditions. While traf1a and traf1b single mutants grow normally under nutrient stress, traf1a traf1b (traf1a/b) double mutants, like the autophagy-defective atg mutants, are hypersensitive to starvation. However, unlike atg mutants, traf1a/b mutants are dwarfed, with an extended lifespan. These mutants, like their atg counterparts, have higher levels of salicylic acid, jasmonic acid, and reactive oxygen species than wild type, along with enhanced cell death, as autophagy negatively regulates these pathways. Finally, like atg mutants, traf1a/b plants have altered biotic and abiotic stress responses compared to wild type, including increased resistance to bacterial infection and reduced resistance to necrotrophic fungal infection and hypoxic stress.
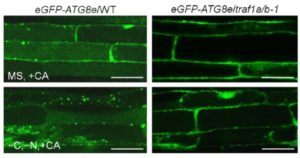 The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.
The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.
Therefore, TRAF1a and TRAF1b regulate autophagy by acting as molecular adapters to help other proteins modulate the stability of ATG6. Many hands make light work.
REFERENCES
Liu, Y., Bassham, D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Ann. Rev. Plant Biol. 63: 215-237.
Qi, H., Xia, F.-N., Xie, L.-N., Yu, L.-N., Chen, Q.-F., Zhuang, X.-H., Wang, Q., Li, F., Jiang, L., Xie, Q., and Xiao, S. (2017). TRAF-Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell 29: doi:10.1015/tpc17.00056.
Wajant, H., Henkler, F., and Scheurich, P. (2001). The TNF-receptor-associated factor family: Scaffold molecules for cytokine receptors, kinases and their regulators. Cellular Signalling 13: 389-400.
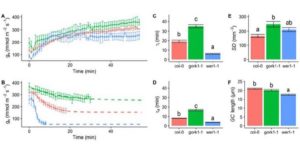 Stomata control gaseous exchange between the leaf and bulk atmosphere limiting CO2 uptake for photosynthesis and water loss by transpiration, and therefore determine plant productivity and water use efficiency. In order to function efficiently, stomata must respond to internal and external signals to balance these two diffusional processes. However, stomatal responses are an order of magnitude slower than photosynthetic responses, which lead to a disconnection between gs and A. Here we discuss the influence of anatomical features on the rapidity of stomatal movement, and explore the temporal relationship between A and gs responses. We describe how these mechanisms have been included into recent modelling efforts, increasing the accuracy and predictive power under dynamic environmental conditions, such as those experienced in the field.
Stomata control gaseous exchange between the leaf and bulk atmosphere limiting CO2 uptake for photosynthesis and water loss by transpiration, and therefore determine plant productivity and water use efficiency. In order to function efficiently, stomata must respond to internal and external signals to balance these two diffusional processes. However, stomatal responses are an order of magnitude slower than photosynthetic responses, which lead to a disconnection between gs and A. Here we discuss the influence of anatomical features on the rapidity of stomatal movement, and explore the temporal relationship between A and gs responses. We describe how these mechanisms have been included into recent modelling efforts, increasing the accuracy and predictive power under dynamic environmental conditions, such as those experienced in the field.

 The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.
The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.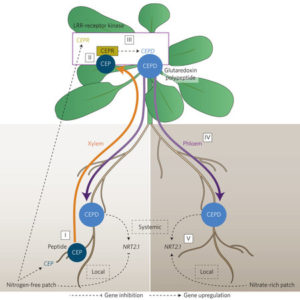 To balance nutrient uptake (usually from heterogeneous sources) with nutrient demand, plants use a root-shoot-root signaling pathway. Previously, a root-to-shoot mobile peptide C-TERMINALLY ENCODED PEPTIDE (CEP) was shown to translocate from N-starved roots to the shoot, where it interacts with a leucine-rich repeat receptor kinase, CEP Receptor 1 (CEPR1). Ohkubo et al. identified two peptides, CEP DOWNSTREAM1 (CEPD1) and CEPD2, that act downstream of CEP-activated CEPR1 and that translocate from shoot to root. CEPD1/2 induce expression of the nitrate transporter NRT2.1 only in roots growing in the presence of nitrate. Therefore, N-starved roots are able to induce nitrate uptake in other roots via this root-shoot-root signalling module: Root-localized CEP sensor detects local N-starvation, ascends to the shoot as a signal, where it induces expression and action of CEPD polypeptides that then translocate to all roots, signaling N demand and resulting in the induction of N-uptake. (Summary by
To balance nutrient uptake (usually from heterogeneous sources) with nutrient demand, plants use a root-shoot-root signaling pathway. Previously, a root-to-shoot mobile peptide C-TERMINALLY ENCODED PEPTIDE (CEP) was shown to translocate from N-starved roots to the shoot, where it interacts with a leucine-rich repeat receptor kinase, CEP Receptor 1 (CEPR1). Ohkubo et al. identified two peptides, CEP DOWNSTREAM1 (CEPD1) and CEPD2, that act downstream of CEP-activated CEPR1 and that translocate from shoot to root. CEPD1/2 induce expression of the nitrate transporter NRT2.1 only in roots growing in the presence of nitrate. Therefore, N-starved roots are able to induce nitrate uptake in other roots via this root-shoot-root signalling module: Root-localized CEP sensor detects local N-starvation, ascends to the shoot as a signal, where it induces expression and action of CEPD polypeptides that then translocate to all roots, signaling N demand and resulting in the induction of N-uptake. (Summary by 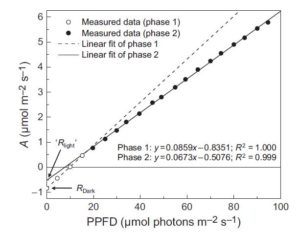 When the uptake of CO2 (A) is plotted against absorbed irradiance (I), at low I there is a noticeable bend that occurs around the light compensation point (I where CO2 release due to mitochondrial respiration is balanced by CO2 uptake by photosynthesis). When the slopes of both parts of the curve are extrapolated back to zero irradiance, it appears as though the rate of mitochondrial respiration is greater in the dark than the light; the inference that there is a light-dependent inhibition of respiration is known as the “Kok effect”. Farquhar and Busch propose a model to explain the Kok effect, in which the apparent curvature of the A vs I plot is attributable to changes in chloroplastic CO2 partial pressure at low I. They suggest that the Kok effect is related to photorespiration, which is consistent with a reduced Kok effect seen in C4 plants, or high CO2 or low O2 conditions. New Phytol.
When the uptake of CO2 (A) is plotted against absorbed irradiance (I), at low I there is a noticeable bend that occurs around the light compensation point (I where CO2 release due to mitochondrial respiration is balanced by CO2 uptake by photosynthesis). When the slopes of both parts of the curve are extrapolated back to zero irradiance, it appears as though the rate of mitochondrial respiration is greater in the dark than the light; the inference that there is a light-dependent inhibition of respiration is known as the “Kok effect”. Farquhar and Busch propose a model to explain the Kok effect, in which the apparent curvature of the A vs I plot is attributable to changes in chloroplastic CO2 partial pressure at low I. They suggest that the Kok effect is related to photorespiration, which is consistent with a reduced Kok effect seen in C4 plants, or high CO2 or low O2 conditions. New Phytol.  Steroids play a role as essential hormones in plants as well as in animals. In plants, steroids termed brassinosteroids (BR) regulate plant growth and development. BR has been shown to be involved in many processes such as light response, stomata and root development, and flowering in plants. BR-regulated gene expression is controlled by the master regulator bri1-EMS-SUPRESSOR1 (BES1). Espinosa-Ruiz et al. show regulation of organ boundary formation in shoot apical meristem and root quiescent center (QC) quiescence by co-repressors BES1 and TOPLESS. BES1 directs TOPLESS to promoters of their target genes (CUC3 and BRAVO) to suppress their expression. (Summary by
Steroids play a role as essential hormones in plants as well as in animals. In plants, steroids termed brassinosteroids (BR) regulate plant growth and development. BR has been shown to be involved in many processes such as light response, stomata and root development, and flowering in plants. BR-regulated gene expression is controlled by the master regulator bri1-EMS-SUPRESSOR1 (BES1). Espinosa-Ruiz et al. show regulation of organ boundary formation in shoot apical meristem and root quiescent center (QC) quiescence by co-repressors BES1 and TOPLESS. BES1 directs TOPLESS to promoters of their target genes (CUC3 and BRAVO) to suppress their expression. (Summary by 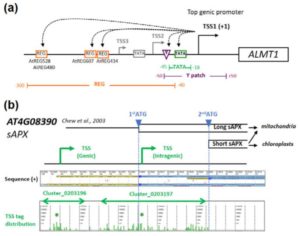 A promoter is a region that “determines the position, direction, frequency, and timing of transcription”. A cell can decode the sequence of the promoter to ensure appropriate transcription, but we still can’t. Tokizawa et al. performed a large-scale survey of promoters by sequencing regions upstream of transcription start sites. They identified promoters corresponding to 80% of Arabidopsis protein-coding genes, as well as many non-coding RNA (miRNA and ncRNA) gene promoters, and intragenic, antisense, and orphan promoters. Onto each of these they mapped core elements (e.g., TATA box) and position-sensitive regulatory elements (REGs). In addition to providing a valuable resource, key findings include that genes average three promoters, and that genes encoding organelle-localized proteins often lack TATA boxes in spite of high expression levels. Plant J.
A promoter is a region that “determines the position, direction, frequency, and timing of transcription”. A cell can decode the sequence of the promoter to ensure appropriate transcription, but we still can’t. Tokizawa et al. performed a large-scale survey of promoters by sequencing regions upstream of transcription start sites. They identified promoters corresponding to 80% of Arabidopsis protein-coding genes, as well as many non-coding RNA (miRNA and ncRNA) gene promoters, and intragenic, antisense, and orphan promoters. Onto each of these they mapped core elements (e.g., TATA box) and position-sensitive regulatory elements (REGs). In addition to providing a valuable resource, key findings include that genes average three promoters, and that genes encoding organelle-localized proteins often lack TATA boxes in spite of high expression levels. Plant J.  Many legumes are important crops, and to date ten legume genomes have been sequenced, including soybean, common bean, mung bean, and two species of wild peanut. Wang et al. used hierarchical comparative genomics analysis of the ten legume genomes, which enabled them to detected gene colinearity between and within genomes, identify colinearity-supported homologs, orthologs and paralogs, relate duplicated genes to specific ancient polyploidization, and characterize the distribution, expansion, and copy-number variations of economically or agriculturally important gene families and regulatory pathways. Additional data and analyses are available at
Many legumes are important crops, and to date ten legume genomes have been sequenced, including soybean, common bean, mung bean, and two species of wild peanut. Wang et al. used hierarchical comparative genomics analysis of the ten legume genomes, which enabled them to detected gene colinearity between and within genomes, identify colinearity-supported homologs, orthologs and paralogs, relate duplicated genes to specific ancient polyploidization, and characterize the distribution, expansion, and copy-number variations of economically or agriculturally important gene families and regulatory pathways. Additional data and analyses are available at Trehalose 6-phosphate (T6P) is a disaccharide formed from two glucose sugars, and more importantly is a signal of glucose availability and regulator of energy homeostasis. Acting via the protein kinase SnRK1, T6P controls the allocation of carbon, leading the plant down a “feast” (growth) or “famine” (survival) metabolic and growth pathway. Paul et al. review the evidence from three cereal crops that altering T6P levels (through GM, marker-assisted selection and novel chemistry) can promote food security through source-sink optimization, for example by stimulating grain filling even under drought conditions. J. Exp. Bot.
Trehalose 6-phosphate (T6P) is a disaccharide formed from two glucose sugars, and more importantly is a signal of glucose availability and regulator of energy homeostasis. Acting via the protein kinase SnRK1, T6P controls the allocation of carbon, leading the plant down a “feast” (growth) or “famine” (survival) metabolic and growth pathway. Paul et al. review the evidence from three cereal crops that altering T6P levels (through GM, marker-assisted selection and novel chemistry) can promote food security through source-sink optimization, for example by stimulating grain filling even under drought conditions. J. Exp. Bot.