Family Chores: TRAF-Family Proteins Help Recycle Cellular Rubbish by Regulating Autophagy Dynamics
IN BRIEF by Jennifer Lockhart [email protected]
Plant cell components that are no longer needed are degraded in the vacuole, but they don’t get there by magic. Sack-like double-membrane structures called autophagosomes engulf this cellular rubbish and neatly transport it to the vacuole for degradation. Autophagy, a highly conserved process orchestrated by a suite of evolutionarily conserved autophagy-related proteins (ATGs), goes into high gear under stress conditions to quickly recycle nutrients and maintain cellular homeostasis (reviewed in Liu and Bassham, 2012). In mammals, upon autophagy induction, the scaffold protein Beclin-1 becomes ubiquitinated by E3 ligases such as Tumor necrosis factor Receptor-Associated Factor (TRAF)-family proteins. Ubiquitinated Beclin-1 stimulates the kinase activity of VPS34, thereby generating the phospholipid PI3P, which acts as a docking site for other components, leading to autophagosome formation. In addition to serving as E3 ligases, TRAF-family proteins play key roles as adaptors in diverse signaling cascades (reviewed in Wajant et al., 2001).
Despite the crucial role of ubiquitination in mammalian autophagosome formation, little is known about this process in plants. Prompted by the notion that TRAF-family proteins might be critical for autophagy in plants as well as mammals, Qi et al. (2017) investigated this family in Arabidopsis thaliana and uncovered an intriguing mechanism for the regulation of plant autophagosome formation. Two proteins, TRAF1a and TRAF1b, localized to the cytoplasm in protoplasts transiently expressing tagged TRAF fusion proteins under normal conditions but to autophagosomes under starvation conditions. While traf1a and traf1b single mutants grow normally under nutrient stress, traf1a traf1b (traf1a/b) double mutants, like the autophagy-defective atg mutants, are hypersensitive to starvation. However, unlike atg mutants, traf1a/b mutants are dwarfed, with an extended lifespan. These mutants, like their atg counterparts, have higher levels of salicylic acid, jasmonic acid, and reactive oxygen species than wild type, along with enhanced cell death, as autophagy negatively regulates these pathways. Finally, like atg mutants, traf1a/b plants have altered biotic and abiotic stress responses compared to wild type, including increased resistance to bacterial infection and reduced resistance to necrotrophic fungal infection and hypoxic stress.
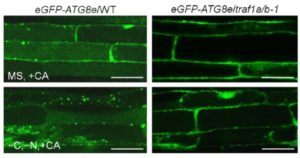 The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.
The authors probed the function of TRAF1a and TRAF1b in autophagy by observing stained autophagosomes in the root cells of wild-type and traf1a/b plants crossed with the autophagosome marker line eGFP-ATG8e treated with the autophagic body stabilizer concanamycin A (CA) to facilitate visualization. Upon starvation, stained vesicles accumulated in wild-type cells but not in the mutant (see figure), indicating that the loss of TRAF1a and TRAF1b prevents starvation-induced autophagosome formation. Both proteins interacted with ATG6, but not with other ATGs, in vitro and in vivo. Since TRAF1a and TRAF1b lack the RING finger domain required for E3 ligase activity, the authors searched for other proteins that interact with ATG6 but have ligase activity. Yeast two-hybrid and co-immunoprecipitation assays indicated that ATG6 interacts with the E3 ligases SINAT1 and SINAT2, which appear to ubiquitinate and destabilize this protein. Importantly, this ubiquitination was strongly reduced in traf1a/b protoplasts compared to wild type, suggesting that TRAF1a and TRAF1b are required for SINAT1- and SINAT2-mediated degradation of ATG6. By contrast, starvation-induced accumulation of SINAT6 reduced this degradation, another process that requires TRAF1a and TRAF1b.
Therefore, TRAF1a and TRAF1b regulate autophagy by acting as molecular adapters to help other proteins modulate the stability of ATG6. Many hands make light work.
REFERENCES
Liu, Y., Bassham, D.C. (2012). Autophagy: Pathways for self-eating in plant cells. Ann. Rev. Plant Biol. 63: 215-237.
Qi, H., Xia, F.-N., Xie, L.-N., Yu, L.-N., Chen, Q.-F., Zhuang, X.-H., Wang, Q., Li, F., Jiang, L., Xie, Q., and Xiao, S. (2017). TRAF-Family Proteins Regulate Autophagy Dynamics by Modulating AUTOPHAGY PROTEIN6 Stability in Arabidopsis. Plant Cell 29: doi:10.1015/tpc17.00056.
Wajant, H., Henkler, F., and Scheurich, P. (2001). The TNF-receptor-associated factor family: Scaffold molecules for cytokine receptors, kinases and their regulators. Cellular Signalling 13: 389-400.







Leave a Reply
Want to join the discussion?Feel free to contribute!