14-3-3 Proteins Function in Plant Immunity
Dong et al. investigate the role of 14-3-3 proteins in plant immunity in Arabidopsis.
By Xiaojing Dong and Jian-Min Zhou
Institute of Genetics and Developmental Biology, Chinese Academy of Sciences, Beijing 100101, China.
Background: The plant innate immune system detects the presence of microbial pathogens and triggers defense responses to terminate or restrict pathogen progression. The BIK1 family receptor-like cytoplasmic kinases (RLCKs) play a crucial role in immune signal transduction and regulate multiple substrate proteins. Much less is known concerning how these kinases and their substrates are organized in a way for robust signaling.
Question: We were interested in identifying interactors of RLCKs and understanding how they regulate immunity.
Findings: We found that the 14-3-3 proteins GRF6 and GRF8 play a key role in both anti-bacterial and anti-fungal immunity in Arabidopsis. We show that these 14-3-3 proteins directly interact with MAPKKK5 to enable their activation by upstream RLCKs. Our study reveals how GRF6 and GRF8 act as a scaffold to regulate the RLCK-MAPKKK5 module to facilitate the activation of MAP kinase cascades during immune signaling.
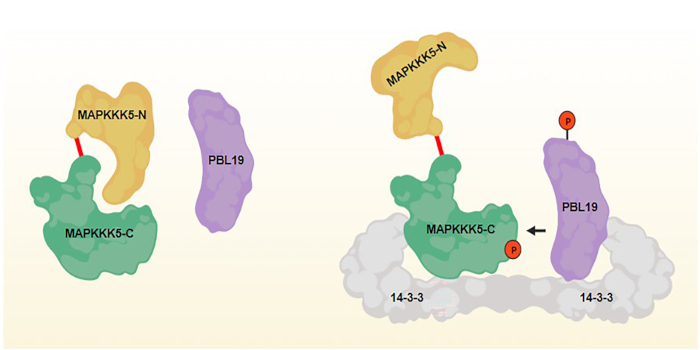
Next steps: It would be of interest to investigate whether different 14-3-3 members regulate different immune responses and how.
Reference:
Shaofei Rao, Zhaoyang Zhou, Pei Miao, Guozhi Bi, Man Hu, Ying Wu, Feng Feng, Xiaojuan Zhang, and Jian-Min Zhou (2018) Roles of receptor-like cytoplasmic Kinase VII members in pattern-triggered immune signaling. Plant Physiol. https://doi.org/10.1104/pp.18.00486
Xiaojing Dong, Feng Feng, Yangjun Li, Lin Li, She Chen, Jian-Min Zhou. (2023) 14-3-3 proteins facilitate the activation of MAP kinase cascades by upstream immunity-related kinases. Plant Cell. https://doi.org/10.1093/plcell/koad088



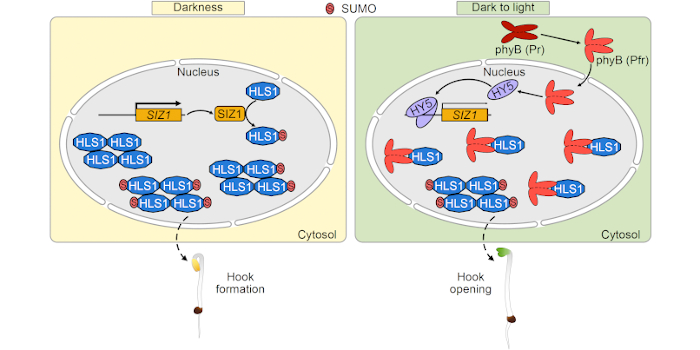 Next steps: We will further study other post-translational modifications of HLS1 in response to light, as well as the effects of SUMOylation on other biochemical functions of HLS1.
Next steps: We will further study other post-translational modifications of HLS1 in response to light, as well as the effects of SUMOylation on other biochemical functions of HLS1.