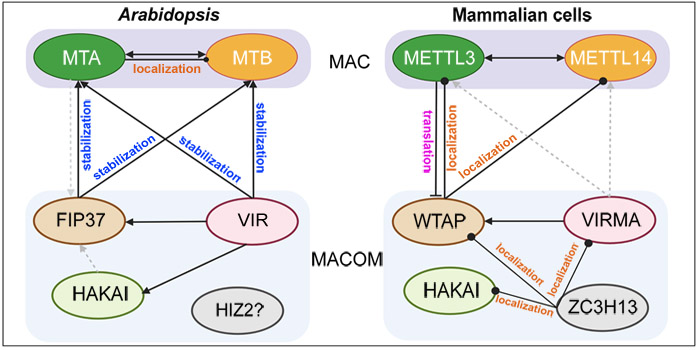
Sending out a salt-related SOS: transcriptional regulation of salt stress responses
Lu et al. explore the roles of a Mediator complex subunit and a WRKY transcription factor in regulating the salt-responsive gene SOS1.
https://doi.org/10.1093/plcell/koad105
Background: Soil salinization is a major environmental hazard that severely affects plant growth and development. Plants have evolved sophisticated mechanisms that help them withstand elevated soil salinity, and Salt Overly Sensitive1 (SOS1) plays a crucial role in plant salt stress tolerance by facilitating the extrusion of excess Na+ from the cells. Although previous reports have demonstrated the important role of post-translational regulation of SOS1 in plant salt stress tolerance, how SOS1 transcription is dynamically modulated in response to different salinity conditions remains unclear.
Question: What are the molecular mechanisms by which plants regulate SOS1 expression at the transcriptional level in response to salinity stress?
Findings: Disruption of the CycC1;1 subunit of the plant Mediator complex promotes salt-induced SOS1 expression and salt tolerance in Arabidopsis because CycC1;1 interferes with RNA polymerase II recruitment by occupying the SOS1 promoter. SOS1 mutation in the cycc1;1 mutant completely compromised its enhanced salt tolerance. Moreover, CycC1;1 can physically interact with the transcription factor WRKY75, which can directly bind to the SOS1 promoter and activate its expression. In contrast to the cycc1;1 mutant, the wrky75 mutant has attenuated SOS1 expression and salt tolerance, whereas overexpression of SOS1 can rescue the salt sensitivity of the mutant. Intriguingly, CycC1;1 inhibits WRKY75 transcriptional activation activity for SOS1 through their interaction; thus, increased SOS1 expression and salt tolerance in the cycc1;1 mutant was abolished by the WRKY75 mutation. In addition, CycC1;1 expression is repressed and WRKY75 expression is stimulated in response to high salinity.
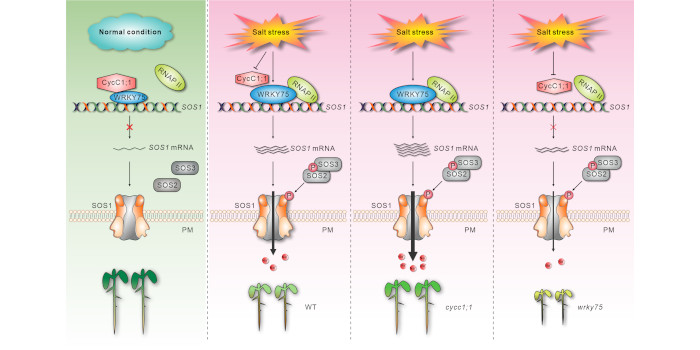
Next steps: In a future study, we will explore whether other components of the Mediator complex are coordinated with CycC1;1 to precisely control SOS1 transcription, and investigate how salinity affects CycC1;1 and WRKY75 expression in the plant’s response to different salinity co
Reference:
Kai-Kai Lu, Ru-Feng Song, Jia-Xing Guo, Yu Zhang, Jia-Xin Zuo, Hui-Hui Chen, Cai-Yi Liao, Xiao-Yu Hu, Feng Ren, Ying-Tang Lu and Wen-Cheng Liu (2023) CycC1;1-WRKY75 complex-mediated transcriptional regulation of SOS1 controls salt stress tolerance in Arabidopsis. https://doi.org/10.1093/plcell/koad105