Review: Molecular concepts to explain heterosis in crops
 Heterosis, the phenomenon where hybrid plants outperform their genetically distinct parents, is a cornerstone of modern agriculture. This review by Hochholdinger and Yu explores the molecular mechanisms behind heterosis. The review traces the history of heterosis, from its early discovery in tobacco by Kölreuter to its widespread use in crops like maize and rice. It covers classical genetic models of heterosis, including dominance, where superior dominant alleles mask recessive ones; overdominance, where heterozygous gene combinations outperform both homozygous forms; and epistasis, where interactions between multiple genes enhance hybrid traits, while integrating modern molecular insights into gene expression, epigenetic modifications, and protein regulation. The review discusses how significant genomic variation between parental lines drives genetic complementation in hybrid plants. This results in the activation of hundreds of additional genes, a process known as single-parent expression (SPE). Additionally, single-gene overdominance plays a significant role, where specific genes in hybrids enhance their vigor. The plant microbiome emerges as a critical factor in hybrid performance. Hybrids and their parental lines differ in microbial communities, particularly in the roots and leaves, influencing traits such as nutrient uptake and stress resistance. Beneficial soil microbes, acting as an extension of the plant genome, further enhance the growth and productivity of hybrids. One insight is the identification of metabolites as biomarkers for predicting heterosis. These metabolites, which fluctuate during critical growth stages, are indicators of traits like biomass and yield. Studies show that metabolite profiles can accurately predict hybrid performance across different environments, offering a powerful tool for breeders to optimize crop yields. (Summary by Amarachi Ezeoke) Trends Plant Sci 10.1016/j.tplants.2024.07.018
Heterosis, the phenomenon where hybrid plants outperform their genetically distinct parents, is a cornerstone of modern agriculture. This review by Hochholdinger and Yu explores the molecular mechanisms behind heterosis. The review traces the history of heterosis, from its early discovery in tobacco by Kölreuter to its widespread use in crops like maize and rice. It covers classical genetic models of heterosis, including dominance, where superior dominant alleles mask recessive ones; overdominance, where heterozygous gene combinations outperform both homozygous forms; and epistasis, where interactions between multiple genes enhance hybrid traits, while integrating modern molecular insights into gene expression, epigenetic modifications, and protein regulation. The review discusses how significant genomic variation between parental lines drives genetic complementation in hybrid plants. This results in the activation of hundreds of additional genes, a process known as single-parent expression (SPE). Additionally, single-gene overdominance plays a significant role, where specific genes in hybrids enhance their vigor. The plant microbiome emerges as a critical factor in hybrid performance. Hybrids and their parental lines differ in microbial communities, particularly in the roots and leaves, influencing traits such as nutrient uptake and stress resistance. Beneficial soil microbes, acting as an extension of the plant genome, further enhance the growth and productivity of hybrids. One insight is the identification of metabolites as biomarkers for predicting heterosis. These metabolites, which fluctuate during critical growth stages, are indicators of traits like biomass and yield. Studies show that metabolite profiles can accurately predict hybrid performance across different environments, offering a powerful tool for breeders to optimize crop yields. (Summary by Amarachi Ezeoke) Trends Plant Sci 10.1016/j.tplants.2024.07.018
Review: Root growth in response to water stress
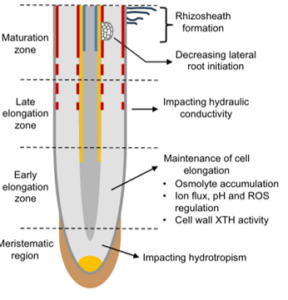 Maintaining root growth is a major plant adaptation to water deficit, enabling continued access to soil water. In a recent review, Voothuluru et al. discuss the inherent complexity of root systems in regard to water stress. Different root types, including primary, seminal, and nodal roots, show varying responses to water deficits, and the root system architecture (RSA) significantly impacts plant performance under water limitation. Growth responses within root growth zones, such as cell production and expansion, determine RSA. Osmotic adjustment is vital for maintaining cell expansion under water stress, with increased solute levels preventing tissue dehydration. Axial water delivery to growth zones and aquaporin-regulated water transport are crucial. Lateral root development, influenced by water availability, shows a biphasic response: promotion under mild deficits and inhibition under severe stress. Interestingly, roots can grow towards water molecules, sometimes defying gravity, in a process called hydrotropism. Because signal sensing, transduction, and root bending involve the entire growth zone and multiple cell types on both sides of the root, further studies with spatial resolution at the tissue- or cell-specific level are needed. The authors observe that improved understanding of root growth will enhance crop productivity under drought. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Cell 10.1093/plcell/koae055
Maintaining root growth is a major plant adaptation to water deficit, enabling continued access to soil water. In a recent review, Voothuluru et al. discuss the inherent complexity of root systems in regard to water stress. Different root types, including primary, seminal, and nodal roots, show varying responses to water deficits, and the root system architecture (RSA) significantly impacts plant performance under water limitation. Growth responses within root growth zones, such as cell production and expansion, determine RSA. Osmotic adjustment is vital for maintaining cell expansion under water stress, with increased solute levels preventing tissue dehydration. Axial water delivery to growth zones and aquaporin-regulated water transport are crucial. Lateral root development, influenced by water availability, shows a biphasic response: promotion under mild deficits and inhibition under severe stress. Interestingly, roots can grow towards water molecules, sometimes defying gravity, in a process called hydrotropism. Because signal sensing, transduction, and root bending involve the entire growth zone and multiple cell types on both sides of the root, further studies with spatial resolution at the tissue- or cell-specific level are needed. The authors observe that improved understanding of root growth will enhance crop productivity under drought. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Cell 10.1093/plcell/koae055
Review: Genetically modified crops and their multifaceted impact on the environment
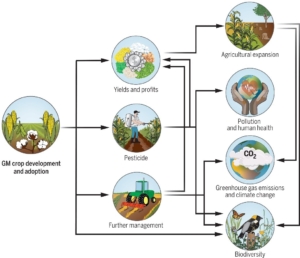 The development of genetically modified (GM) crops aims to improve agricultural yields in the field. However, their incorporation into agricultural systems is complex, as regulations and acceptance vary globally. While some countries embrace GM crops with herbicide and insect resistance traits, others remain cautious due to concerns about environmental and societal impacts. A recent review by Noack et al. in Science explores both the direct and indirect consequences of GM crop adoption on the environment, including effects on deforestation, human health, pesticide use, and biodiversity. The authors specifically consider the environmental impacts in both adopting and non-adopting countries to provide a global perspective. They note that while GM crops often increase yields, their effects on other environmental factors are complex and context-dependent. A major determinant of their environmental impact is how they are integrated into broader agricultural systems. The authors also highlight the limitations of current studies, which mainly focus on two traits: herbicide tolerance and insect resistance. Broader adoption of GM crops with traits like abiotic stress tolerance could present clearer environmental benefits. Lastly, this review underscores the complexity of GM crop adoption on a global scale and highlights the need for powerful modelling tools for a realistic assessment of their impact on the environment. These could offer the necessary scientific data to make informed decisions on the adoption of GM crops to combat climate change. (Summary by Thomas Depaepe @thdpaepe). Science 10.1126/science.ado934
The development of genetically modified (GM) crops aims to improve agricultural yields in the field. However, their incorporation into agricultural systems is complex, as regulations and acceptance vary globally. While some countries embrace GM crops with herbicide and insect resistance traits, others remain cautious due to concerns about environmental and societal impacts. A recent review by Noack et al. in Science explores both the direct and indirect consequences of GM crop adoption on the environment, including effects on deforestation, human health, pesticide use, and biodiversity. The authors specifically consider the environmental impacts in both adopting and non-adopting countries to provide a global perspective. They note that while GM crops often increase yields, their effects on other environmental factors are complex and context-dependent. A major determinant of their environmental impact is how they are integrated into broader agricultural systems. The authors also highlight the limitations of current studies, which mainly focus on two traits: herbicide tolerance and insect resistance. Broader adoption of GM crops with traits like abiotic stress tolerance could present clearer environmental benefits. Lastly, this review underscores the complexity of GM crop adoption on a global scale and highlights the need for powerful modelling tools for a realistic assessment of their impact on the environment. These could offer the necessary scientific data to make informed decisions on the adoption of GM crops to combat climate change. (Summary by Thomas Depaepe @thdpaepe). Science 10.1126/science.ado934
Timekeeping mechanism in prokaryotes: Cyanobacteria can anticipate the seasons
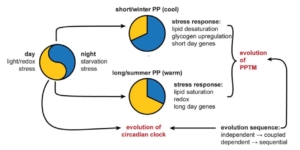 Cyanobacteria, although short-lived, are sensitive to photoperiodic time measurement just as in eukaryotes. Jabbur et al. discuss the discovery of photoperiodism in prokaryotic cyanobacteria, Synechococcus elongatus and their ability to anticipate seasonal changes through light and dark cycles similar to eukaryotes. This ability is linked to their circadian clock, which helps them to respond to physiological changes, such as resistance to cold, depending on prior exposure to day length. S. elongatus was discovered to develop increased resistance to cold when exposed to shorter winter-like days, while this response is absent in mutant strains lacking the circadian clock genes. RNA-seq analysis further revealed that the short-day exposure enhances lipid desaturation, which is an important adaptation for survival in cold conditions, while exposure to long days activates heat and light stress responses. This suggests that cyanobacteria possibly evolved photoperiodic time measurement from stress response mechanisms, helping to prepare them for seasonal changes and adapt to their environment. This challenges the traditional belief that only higher organisms could anticipate seasonal changes. Understanding photoperiodic responses in cyanobacteria offers a simple model to study mechanisms of photoperiodism and can also provide insight into how they form harmful blooms, which have significant ecological impact. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Science 10.1126/science.ado8588
Cyanobacteria, although short-lived, are sensitive to photoperiodic time measurement just as in eukaryotes. Jabbur et al. discuss the discovery of photoperiodism in prokaryotic cyanobacteria, Synechococcus elongatus and their ability to anticipate seasonal changes through light and dark cycles similar to eukaryotes. This ability is linked to their circadian clock, which helps them to respond to physiological changes, such as resistance to cold, depending on prior exposure to day length. S. elongatus was discovered to develop increased resistance to cold when exposed to shorter winter-like days, while this response is absent in mutant strains lacking the circadian clock genes. RNA-seq analysis further revealed that the short-day exposure enhances lipid desaturation, which is an important adaptation for survival in cold conditions, while exposure to long days activates heat and light stress responses. This suggests that cyanobacteria possibly evolved photoperiodic time measurement from stress response mechanisms, helping to prepare them for seasonal changes and adapt to their environment. This challenges the traditional belief that only higher organisms could anticipate seasonal changes. Understanding photoperiodic responses in cyanobacteria offers a simple model to study mechanisms of photoperiodism and can also provide insight into how they form harmful blooms, which have significant ecological impact. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Science 10.1126/science.ado8588
Single cell transcriptomics after mild drought reveals two types of mesophyll responses
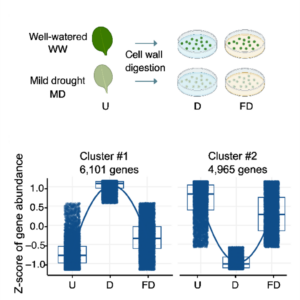 Single-cell transcriptomics offers unprecedented insights into how plants respond to their environment. Although this technique is powerful, it requires extensive processing of the tissue (cell wall digestion to form protoplasts) prior to sequencing, which can induce additional transcriptional changes. Here, Tenorio Berrío et al. incorporated into their protocol a block on transcription through applying actinomycin D (ActD) prior to cell isolation. Using this method, they investigated gene expression in young Arabidopsis leaves following mild drought. They compared these new results to those from a prior study they had conducted without the transcriptional block and found significant differences in gene expression. Interestingly, many known drought-response genes were absent from the samples that had been digested without the ActD, suggesting that the transcriptional response to cell wall degradation masked the transcriptional response to drought. They followed this up with whole-mount in situ hybridization studies of several genes. From the dataset obtained from the ActD treated sample, they identified two subpopulations of mesophyll cells as defined by their transcriptomes. One, located around the leaf margin and near the veins, is characterized by drought-responsive genes. A spatially distinct set, closer to the main photosynthetic cells, showed an upregulation of iron starvation-responsive genes; the authors discuss the potential origin of this response. The article illustrates the value of using ActD to arrest transcription prior to single cell isolation. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.30.61043
Single-cell transcriptomics offers unprecedented insights into how plants respond to their environment. Although this technique is powerful, it requires extensive processing of the tissue (cell wall digestion to form protoplasts) prior to sequencing, which can induce additional transcriptional changes. Here, Tenorio Berrío et al. incorporated into their protocol a block on transcription through applying actinomycin D (ActD) prior to cell isolation. Using this method, they investigated gene expression in young Arabidopsis leaves following mild drought. They compared these new results to those from a prior study they had conducted without the transcriptional block and found significant differences in gene expression. Interestingly, many known drought-response genes were absent from the samples that had been digested without the ActD, suggesting that the transcriptional response to cell wall degradation masked the transcriptional response to drought. They followed this up with whole-mount in situ hybridization studies of several genes. From the dataset obtained from the ActD treated sample, they identified two subpopulations of mesophyll cells as defined by their transcriptomes. One, located around the leaf margin and near the veins, is characterized by drought-responsive genes. A spatially distinct set, closer to the main photosynthetic cells, showed an upregulation of iron starvation-responsive genes; the authors discuss the potential origin of this response. The article illustrates the value of using ActD to arrest transcription prior to single cell isolation. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.30.61043
Zinc regulates symbiotic nitrogen fixation in response to soil nitrate
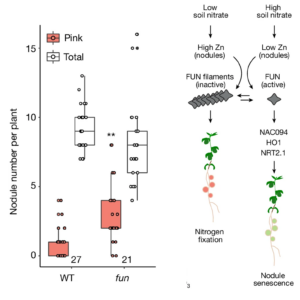 Legume plants inhibit the energy-intensive process of symbiotic nitrogen fixation when sufficient nitrate is present, primarily through nodule senescence. However, the nodule-specific genetic regulatory mechanisms of nitrate sensing and nodule senescence are poorly understood. A recent study by Lin and Bjørk et al. identified a key genetic regulator. The authors conducted a large-scale genetic screening from a population of insertional mutants of the nodule-forming legume lotus and identified a mutant that maintained nodules even in the presence of ample nitrogen. The affected gene in the mutant, termed FUN (fixation under nitrate), encodes a protein that acts as a nodule-specific transcriptional factor that positively regulates nodule senescence under high nitrate. Notably, FUN is regulated at the protein level through its zinc-binding sensor domain. When Zn levels increase, FUN transitions from an active monomeric state to inactive filamentous form, inhibiting the transcription of downstream genes associated with nodule senescence. Using zinc sensitive dye, X-ray fluorescence, and reporter lines, the authors established an inverse relationship between nitrate availability and intracellular zinc concentration in nodules, confirming zinc’s role as a secondary messenger. This study provides new insights into the regulation of nitrogen fixation in legumes. (Summary by Arijit Mukherjee @ArijitM61745830) Nature 10.1038/s41586-024-07607-6
Legume plants inhibit the energy-intensive process of symbiotic nitrogen fixation when sufficient nitrate is present, primarily through nodule senescence. However, the nodule-specific genetic regulatory mechanisms of nitrate sensing and nodule senescence are poorly understood. A recent study by Lin and Bjørk et al. identified a key genetic regulator. The authors conducted a large-scale genetic screening from a population of insertional mutants of the nodule-forming legume lotus and identified a mutant that maintained nodules even in the presence of ample nitrogen. The affected gene in the mutant, termed FUN (fixation under nitrate), encodes a protein that acts as a nodule-specific transcriptional factor that positively regulates nodule senescence under high nitrate. Notably, FUN is regulated at the protein level through its zinc-binding sensor domain. When Zn levels increase, FUN transitions from an active monomeric state to inactive filamentous form, inhibiting the transcription of downstream genes associated with nodule senescence. Using zinc sensitive dye, X-ray fluorescence, and reporter lines, the authors established an inverse relationship between nitrate availability and intracellular zinc concentration in nodules, confirming zinc’s role as a secondary messenger. This study provides new insights into the regulation of nitrogen fixation in legumes. (Summary by Arijit Mukherjee @ArijitM61745830) Nature 10.1038/s41586-024-07607-6
Pressed for time: Why do herbarium collections still matter in a digital world?
 For centuries, herbarium collections have provided critical data about plant species and their abundance. However, with the development of digital apps such as iNaturalist that allow citizen scientists to enter data about where plants are found, questions arise about whether herbarium collections are still relevant. Eckert et al. studied how herbarium and iNaturalist data influence the understanding of vascular plant diversity in Canada. Without doubt, herbarium collections are still very important because they provide real evidence for identifying plants, they keep track of how the distribution of species has changed over time, and they help with conservation by keeping rare and endangered specimens secure. This study used phylogenetic and functional trait analyses from the TRY database (TRY is a sentiment, not an acromym) to evaluate the completeness of both herbarium and iNaturalist records. Significant functional and taxonomic voids were identified, particularly in herbarium data, which frequently lacked comprehensive species coverage. The use of iNaturalist data as a supplement to herbarium records provides a more diverse understanding of plant species distributions and features, which is critical for improving the quality and comprehensiveness of plant diversity assessments. The study emphasizes the potential benefits of digitizing the remaining 7.3 million herbarium specimens to overcome these gaps and increase the overall representation of plant variety. (Summary by Tuyelee Das @das_tuyelee) Nature Comms. 10.1038/s41467-024-51899-1
For centuries, herbarium collections have provided critical data about plant species and their abundance. However, with the development of digital apps such as iNaturalist that allow citizen scientists to enter data about where plants are found, questions arise about whether herbarium collections are still relevant. Eckert et al. studied how herbarium and iNaturalist data influence the understanding of vascular plant diversity in Canada. Without doubt, herbarium collections are still very important because they provide real evidence for identifying plants, they keep track of how the distribution of species has changed over time, and they help with conservation by keeping rare and endangered specimens secure. This study used phylogenetic and functional trait analyses from the TRY database (TRY is a sentiment, not an acromym) to evaluate the completeness of both herbarium and iNaturalist records. Significant functional and taxonomic voids were identified, particularly in herbarium data, which frequently lacked comprehensive species coverage. The use of iNaturalist data as a supplement to herbarium records provides a more diverse understanding of plant species distributions and features, which is critical for improving the quality and comprehensiveness of plant diversity assessments. The study emphasizes the potential benefits of digitizing the remaining 7.3 million herbarium specimens to overcome these gaps and increase the overall representation of plant variety. (Summary by Tuyelee Das @das_tuyelee) Nature Comms. 10.1038/s41467-024-51899-1
Comment: Lack of racial diversity in UK plant science
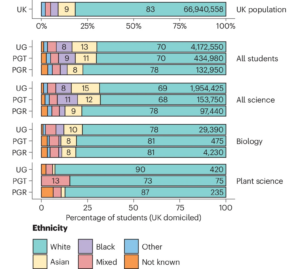 In most STEM disciplines there is a racial disparity between practitioners and the broader population. This disparity is often greater in plant sciences than in biomedical-related disciplines. Here, Hubbard and colleagues take a close look at the demographics of plant scientists in the UK and provide some recommendations for action. Contributing factors could include where plant science is taught (not in London, and in highly selective institutions) as well as disparity in overall admissions in higher education between white and Black and Asian students. The authors also point to the fact that Black and Asian families are less likely to have gardens than white families, contributing to a lack of plant awareness amongst these students. The authors discuss the impacts of racism and colonialism underlying botanical history, including that much of the labor done by enslaved people was crop cultivation and harvesting. They provide a thoughtful list of recommendations, including increased recruitment and representation, but also the need for teachers and mentors to be aware of how their actions can contribute to a hostile, “othering” climate. Please read it and think about how you can apply these approaches in your institution, teaching, mentoring and outreach efforts. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01778-w
In most STEM disciplines there is a racial disparity between practitioners and the broader population. This disparity is often greater in plant sciences than in biomedical-related disciplines. Here, Hubbard and colleagues take a close look at the demographics of plant scientists in the UK and provide some recommendations for action. Contributing factors could include where plant science is taught (not in London, and in highly selective institutions) as well as disparity in overall admissions in higher education between white and Black and Asian students. The authors also point to the fact that Black and Asian families are less likely to have gardens than white families, contributing to a lack of plant awareness amongst these students. The authors discuss the impacts of racism and colonialism underlying botanical history, including that much of the labor done by enslaved people was crop cultivation and harvesting. They provide a thoughtful list of recommendations, including increased recruitment and representation, but also the need for teachers and mentors to be aware of how their actions can contribute to a hostile, “othering” climate. Please read it and think about how you can apply these approaches in your institution, teaching, mentoring and outreach efforts. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01778-w
 In most STEM disciplines there is a racial disparity between practitioners and the broader population. This disparity is often greater in plant sciences than in biomedical-related disciplines. Here, Hubbard and colleagues take a close look at the demographics of plant scientists in the UK and provide some recommendations for action. Contributing factors could include where plant science is taught (not in London, and in highly selective institutions) as well as disparity in overall admissions in higher education between white and Black and Asian students. The authors also point to the fact that Black and Asian families are less likely to have gardens than white families, contributing to a lack of plant awareness amongst these students. The authors discuss the impacts racism and colonialism underlying botanical history, including that much of the labor done by enslaved people was crop cultivation and harvesting. They provide a thoughtful list of recommendations, including increased recruitment and representation, but also the need for teachers and mentors to be aware of how their actions can contribute to a hostile, “othering” climate. Please read it and think about how you can apply these approaches in your institution, teaching, mentoring and outreach efforts. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01778-w
In most STEM disciplines there is a racial disparity between practitioners and the broader population. This disparity is often greater in plant sciences than in biomedical-related disciplines. Here, Hubbard and colleagues take a close look at the demographics of plant scientists in the UK and provide some recommendations for action. Contributing factors could include where plant science is taught (not in London, and in highly selective institutions) as well as disparity in overall admissions in higher education between white and Black and Asian students. The authors also point to the fact that Black and Asian families are less likely to have gardens than white families, contributing to a lack of plant awareness amongst these students. The authors discuss the impacts racism and colonialism underlying botanical history, including that much of the labor done by enslaved people was crop cultivation and harvesting. They provide a thoughtful list of recommendations, including increased recruitment and representation, but also the need for teachers and mentors to be aware of how their actions can contribute to a hostile, “othering” climate. Please read it and think about how you can apply these approaches in your institution, teaching, mentoring and outreach efforts. (Summary by Mary Williams @PlantTeaching) Nature Plants 10.1038/s41477-024-01778-w