Review. Chilling out: How plants remodel membranes to survive the cold
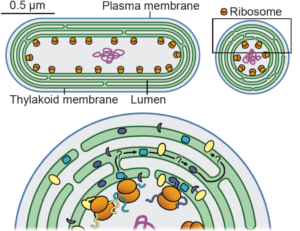
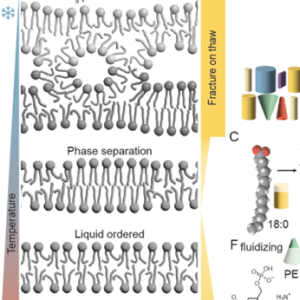 Low temperatures disrupt cellular processes and require metabolic adaptation. Cold-induced lipid remodeling helps maintain membrane permeability and prevents phase separations that can lead to cellular damage. In this review, Shomo, Li and colleagues provide an overview of the mechanisms by which plants remodel their membrane lipids to adapt to low temperatures and cold stress. They focus on glycerolipids, sphingolipids, and phytosterols. Additionally, they examine how cold stress affects membrane composition and functionality at the subcellular level, presenting current glycerolipidomic data from Arabidopsis thaliana, maize, and sorghum. This data highlights the consistency as well as the variability of lipid responses across species. The authors delve into the three main ways of lipid alteration. First is acyl editing and headgroup exchange, where plants modify their membrane lipids to maintain membrane integrity and function. The second way is regulating lipase activity, which plays a crucial role in lipid remodeling during cold stress. The third is changes in phytosterol abundance, where altering phytosterol levels help to stabilize membranes under cold conditions. The review points out recent advances and accomplishments in the field with an informative overview of observed lipid changes, and highlights gaps in current the knowledge and areas needing further investigation to fully understand and manipulate plant cold tolerance. Summary by Ann-Kathrin Rößling (@AK_Roessling) 10.1093/plphys/kiae382
Low temperatures disrupt cellular processes and require metabolic adaptation. Cold-induced lipid remodeling helps maintain membrane permeability and prevents phase separations that can lead to cellular damage. In this review, Shomo, Li and colleagues provide an overview of the mechanisms by which plants remodel their membrane lipids to adapt to low temperatures and cold stress. They focus on glycerolipids, sphingolipids, and phytosterols. Additionally, they examine how cold stress affects membrane composition and functionality at the subcellular level, presenting current glycerolipidomic data from Arabidopsis thaliana, maize, and sorghum. This data highlights the consistency as well as the variability of lipid responses across species. The authors delve into the three main ways of lipid alteration. First is acyl editing and headgroup exchange, where plants modify their membrane lipids to maintain membrane integrity and function. The second way is regulating lipase activity, which plays a crucial role in lipid remodeling during cold stress. The third is changes in phytosterol abundance, where altering phytosterol levels help to stabilize membranes under cold conditions. The review points out recent advances and accomplishments in the field with an informative overview of observed lipid changes, and highlights gaps in current the knowledge and areas needing further investigation to fully understand and manipulate plant cold tolerance. Summary by Ann-Kathrin Rößling (@AK_Roessling) 10.1093/plphys/kiae382
Review: Defining and rewiring of gene regulatory networks for plant improvement
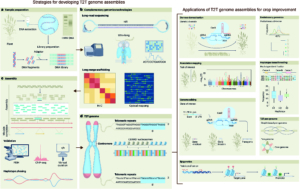
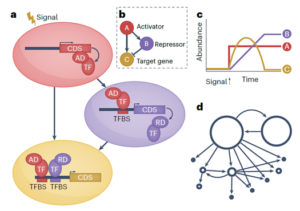 Much of the focus of functional genomics studies in plants is to improve yield, disease resistance, abiotic stress tolerance and nutritional quality. Many desirable traits are controlled by sets of genes that are coordinated in a complex network, called a gene regulatory network (GRN). A transcription factor (TF) binds to the promoter region of one or more genes and promotes or represses gene expression. In this review, Borowsky and Bailey-Serres discuss how defining the GRNs underlying desired traits can be used to fine-tune and further adjust them to meet the demands of climate change and food supply. Next-generation sequencing technologies coupled with omics studies made it possible to explore the gene circuitry and interacting proteins controlling favorable traits. The functions of most major genes are conserved among different species with much diversity attributed to TFs and cis-regulatory elements, which drive or alter the expression of target genes. For example, the function of the root TF SHORTROOT is conserved, but its redeployment results in the addition of more cortical cells in maize compared to Arabidopsis. Similarly, the function of the bundle sheath TF DNA BINDING WITH ONE FINGER is conserved in photosynthesis, however, its downstream deployment regulates C3 genes in rice and C4 genes in sorghum. The authors present a set of examples of how synthetic biology and genome editing can be used to engineer desirable phenotypes in plants through rewiring gene circuitry. (Summary by Asif Ali @pbgasifkalas) Nat. Genet. 10.1038/s41588-024-01806-7
Much of the focus of functional genomics studies in plants is to improve yield, disease resistance, abiotic stress tolerance and nutritional quality. Many desirable traits are controlled by sets of genes that are coordinated in a complex network, called a gene regulatory network (GRN). A transcription factor (TF) binds to the promoter region of one or more genes and promotes or represses gene expression. In this review, Borowsky and Bailey-Serres discuss how defining the GRNs underlying desired traits can be used to fine-tune and further adjust them to meet the demands of climate change and food supply. Next-generation sequencing technologies coupled with omics studies made it possible to explore the gene circuitry and interacting proteins controlling favorable traits. The functions of most major genes are conserved among different species with much diversity attributed to TFs and cis-regulatory elements, which drive or alter the expression of target genes. For example, the function of the root TF SHORTROOT is conserved, but its redeployment results in the addition of more cortical cells in maize compared to Arabidopsis. Similarly, the function of the bundle sheath TF DNA BINDING WITH ONE FINGER is conserved in photosynthesis, however, its downstream deployment regulates C3 genes in rice and C4 genes in sorghum. The authors present a set of examples of how synthetic biology and genome editing can be used to engineer desirable phenotypes in plants through rewiring gene circuitry. (Summary by Asif Ali @pbgasifkalas) Nat. Genet. 10.1038/s41588-024-01806-7
Review: Stem-borne roots as a framework to study trans-organogenesis
 Plants develop new organs and tissues throughout their lifespan as they grow new leaves, roots and reproductive structures. Many of these tissues arise from similar tissues, such as lateral roots arising from primary roots, and the mechanisms guiding their formation are well understood. But what about ‘trans-organogenesis’: where structures arise from completely different tissue types? The development of roots from stems – also known as adventitious roots – is one such example. And while stem-borne roots have been described for many species, the mechanisms controlling their development remain elusive. In this review, Rasmussen et al. summarise the current knowledge on development of stem-borne roots, including their occurrence throughout the plant kingdom and diversity of form and function. They consider open questions of whether stem-borne roots share an evolutionary history, or have arisen multiple times, possibly through rewiring of ancient developmental pathways. The authors provide a framework to better classify types of stem-borne roots based on their form, function and tissue of origin – and propose stem-borne roots as a model for understanding trans-organogenesis, plasticity and adaptation in plants. (Summary by Alicia Quinn @AliciaQuinnSci). Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102604
Plants develop new organs and tissues throughout their lifespan as they grow new leaves, roots and reproductive structures. Many of these tissues arise from similar tissues, such as lateral roots arising from primary roots, and the mechanisms guiding their formation are well understood. But what about ‘trans-organogenesis’: where structures arise from completely different tissue types? The development of roots from stems – also known as adventitious roots – is one such example. And while stem-borne roots have been described for many species, the mechanisms controlling their development remain elusive. In this review, Rasmussen et al. summarise the current knowledge on development of stem-borne roots, including their occurrence throughout the plant kingdom and diversity of form and function. They consider open questions of whether stem-borne roots share an evolutionary history, or have arisen multiple times, possibly through rewiring of ancient developmental pathways. The authors provide a framework to better classify types of stem-borne roots based on their form, function and tissue of origin – and propose stem-borne roots as a model for understanding trans-organogenesis, plasticity and adaptation in plants. (Summary by Alicia Quinn @AliciaQuinnSci). Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102604
Cytokinin signalling regulates auxin availability and wounding-induced adventitious rooting competency
 Adventitious roots, i.e. those derived from non-root tissues, are key elements of the plastic architecture of root systems. Auxins and cytokinins are known to induce and inhibit (respectively) adventitious root formation, though the exact mechanism remains to be fully explained. Here, Damodaran and Strader investigated the role of local conversion of indole-3-butyric acid (IBA) into indole-3-acetic acid (IAA) – both forms of auxin – in adventitious rooting competency. Following wounding of the hypocotyl, adventitious roots form mainly near the excision site, indicating that not all hypocotyl cells are competent to make roots. IAA can be obtained locally from IBA via peroxisomal beta-oxidation, and mutants unable to make this conversion produce fewer adventitious roots than the wildtype. TRANSPORTER OF IBA 1 (TOB1) can limit the supply of IBA by sequestering it into the vacuole, and tob1 plants produced more adventitious roots than wildtype, further evidencing the role of IBA-to-IAA conversion in adventitious rooting competency. Additionally, tob1 hypocotyls were insensitive to the known inhibitory effects of cytokinin on adventitious rooting, possibly due to cytokinin control of TOB1 expression. These results contribute to a more mechanistic understanding of the long-established auxin-cytokinin interplay in adventitious rooting, which will be invaluable in the development of improved protocols for clonal propagation by cuttings. (Summary by John Vilasboa @vilasjohn) Dev. Cell. 10.1016/j.devcel.2024.06.019
Adventitious roots, i.e. those derived from non-root tissues, are key elements of the plastic architecture of root systems. Auxins and cytokinins are known to induce and inhibit (respectively) adventitious root formation, though the exact mechanism remains to be fully explained. Here, Damodaran and Strader investigated the role of local conversion of indole-3-butyric acid (IBA) into indole-3-acetic acid (IAA) – both forms of auxin – in adventitious rooting competency. Following wounding of the hypocotyl, adventitious roots form mainly near the excision site, indicating that not all hypocotyl cells are competent to make roots. IAA can be obtained locally from IBA via peroxisomal beta-oxidation, and mutants unable to make this conversion produce fewer adventitious roots than the wildtype. TRANSPORTER OF IBA 1 (TOB1) can limit the supply of IBA by sequestering it into the vacuole, and tob1 plants produced more adventitious roots than wildtype, further evidencing the role of IBA-to-IAA conversion in adventitious rooting competency. Additionally, tob1 hypocotyls were insensitive to the known inhibitory effects of cytokinin on adventitious rooting, possibly due to cytokinin control of TOB1 expression. These results contribute to a more mechanistic understanding of the long-established auxin-cytokinin interplay in adventitious rooting, which will be invaluable in the development of improved protocols for clonal propagation by cuttings. (Summary by John Vilasboa @vilasjohn) Dev. Cell. 10.1016/j.devcel.2024.06.019
Convergent evolution of plant prickles
 Contrary to common belief, roses do not have thorns: instead, they have prickles. Thorns (as in hawthorns) are modified stems, spines (as in cactus spines) are modified leaves, and prickles (as in roses) are modified epidermal tissues. Prickles occur in a wide range of plants. Satterlee et al. set out to investigate the genetic basis for prickle formation, starting with a comparison between a prickleless cultivated eggplant (Solanum melongena) and its prickled wild relative (Solanum insanum). Interestingly, they identified a gene known to be involved in cytokinin biosynthesis, LONELY GUY (LOG), as critical for prickle formation. The authors looked across the Solanum genus and beyond and found that mutations in LOG are widely correlated with prickle loss. Furthermore, homologous prickle-like structures known as barbs in grasses are also dependent on a functional LOG gene, indicating that loss of prickles has occurred convergently. Finally, the authors used gene editing techniques to produce prickleless varieties without additional pleiotropic effects. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.ado1663
Contrary to common belief, roses do not have thorns: instead, they have prickles. Thorns (as in hawthorns) are modified stems, spines (as in cactus spines) are modified leaves, and prickles (as in roses) are modified epidermal tissues. Prickles occur in a wide range of plants. Satterlee et al. set out to investigate the genetic basis for prickle formation, starting with a comparison between a prickleless cultivated eggplant (Solanum melongena) and its prickled wild relative (Solanum insanum). Interestingly, they identified a gene known to be involved in cytokinin biosynthesis, LONELY GUY (LOG), as critical for prickle formation. The authors looked across the Solanum genus and beyond and found that mutations in LOG are widely correlated with prickle loss. Furthermore, homologous prickle-like structures known as barbs in grasses are also dependent on a functional LOG gene, indicating that loss of prickles has occurred convergently. Finally, the authors used gene editing techniques to produce prickleless varieties without additional pleiotropic effects. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.ado1663
A kinase fusion protein from Aegilops longissima confers resistance to wheat powdery mildew
 The obligate biotrophic pathogen Blumeria graminis f. sp. tritici (Bgt) is the cause of wheat powdery mildew, a foliar disease that results in significant yield loss in most wheat-growing regions across the globe. This research focuses on the identification and functional analysis of the powdery mildew resistance gene Pm13, which was introgressed into wheat from Aegilops longissima. The gene was identified through a comprehensive approach that revealed its critical role in conferring resistance to wheat powdery mildew. Pm13 encodes a unique fusion protein that combines elements typically associated with the regulation and signaling of cell death. The validation of Pm13 function, confirmed through various genetic assays, underscores its importance in enhancing disease resistance. Notably, a specific segment of the protein was shown to initiate cell death, highlighting the gene’s potential impact on improving wheat resilience through targeted breeding or genetic engineering. This discovery adds a valuable tool to the arsenal for combating powdery mildew in wheat. The findings also highlight the potential of wild relatives, such as Aegilops longissima, to enhance disease resistance in cultivated wheat. (Summary by Muhammad Aamir Khan @MAKNature1998) Nature 10.1038/s41467-024-50909-6
The obligate biotrophic pathogen Blumeria graminis f. sp. tritici (Bgt) is the cause of wheat powdery mildew, a foliar disease that results in significant yield loss in most wheat-growing regions across the globe. This research focuses on the identification and functional analysis of the powdery mildew resistance gene Pm13, which was introgressed into wheat from Aegilops longissima. The gene was identified through a comprehensive approach that revealed its critical role in conferring resistance to wheat powdery mildew. Pm13 encodes a unique fusion protein that combines elements typically associated with the regulation and signaling of cell death. The validation of Pm13 function, confirmed through various genetic assays, underscores its importance in enhancing disease resistance. Notably, a specific segment of the protein was shown to initiate cell death, highlighting the gene’s potential impact on improving wheat resilience through targeted breeding or genetic engineering. This discovery adds a valuable tool to the arsenal for combating powdery mildew in wheat. The findings also highlight the potential of wild relatives, such as Aegilops longissima, to enhance disease resistance in cultivated wheat. (Summary by Muhammad Aamir Khan @MAKNature1998) Nature 10.1038/s41467-024-50909-6
New kid on the plant block: Single-cell proteomics
 While single-cell omics technologies, particularly transcriptomics, are already becoming widely adopted in plant science, quantifying proteins at single cell resolution is less established. Fortunately, important technological strides have been made that improve sample preparation, separation techniques, and overall sensitivity and resolution to make single cell proteomics (SCP) possible. Montes et al. have recently developed a multiplexed methodology for SCP specifically adapted to plant tissue, overcoming typical challenges associated with handling such samples. As a proof-of-concept, the authors compared proteins from two adjacent root cell layers, the endodermis and cortex, and observed their distinct proteomic signatures. The optimized SCP can successfully detect proteins associated with both low and high abundance transcripts, and even detects low abundant proteins like transcription factors, opening up tremendous possibilities to study plant signaling networks in neighboring cell types. As expected however, the varying cell sizes along the root led to considerable variation in protein quantification, an important consideration for improvement in future studies. In conclusion, this methodology offers a clear, comprehensive, and practical tool for scientists looking to explore the world of single-cell proteomics in plants. (Summary by Thomas Depaepe @thdpaepe). New Phytologist 10.1111/nph.19923
While single-cell omics technologies, particularly transcriptomics, are already becoming widely adopted in plant science, quantifying proteins at single cell resolution is less established. Fortunately, important technological strides have been made that improve sample preparation, separation techniques, and overall sensitivity and resolution to make single cell proteomics (SCP) possible. Montes et al. have recently developed a multiplexed methodology for SCP specifically adapted to plant tissue, overcoming typical challenges associated with handling such samples. As a proof-of-concept, the authors compared proteins from two adjacent root cell layers, the endodermis and cortex, and observed their distinct proteomic signatures. The optimized SCP can successfully detect proteins associated with both low and high abundance transcripts, and even detects low abundant proteins like transcription factors, opening up tremendous possibilities to study plant signaling networks in neighboring cell types. As expected however, the varying cell sizes along the root led to considerable variation in protein quantification, an important consideration for improvement in future studies. In conclusion, this methodology offers a clear, comprehensive, and practical tool for scientists looking to explore the world of single-cell proteomics in plants. (Summary by Thomas Depaepe @thdpaepe). New Phytologist 10.1111/nph.19923
Unforeseen plant diversity in global dryland
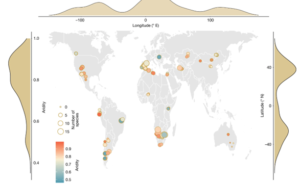 Earth’s terrestrial ecosystems harbor remarkable plant diversity. Some of this diversity can be quantified as functional traits, which provide a measure for diversity in different environments. In this new work, Gross et al. used a functional trait analysis to examine how plants respond to increasing aridity and animal grazing in global drylands, which are highly vulnerable to drivers of global change. The study analyzed 133,769 traits from 301 perennial plants across 98 sites in 25 countries. The findings break the widely accepted belief that extreme climatic conditions lead to a decline in the diversity of plant traits. On the contrary, it suggests that plants in arid regions use a wide variety of adaptation strategies, and this diversity increases with the degree of aridity. The authors describe this response to aridity as the “plant loneliness syndrome”. Heavily grazed drylands also correspond to greater diversity in traits. The findings emphasize that factors like soil fertility and plant cover contribute to trait diversity in challenging environments. This study underscores the significance of drylands as a global reservoir of plant diversity and shows how plants can adapt to increasing environmental pressures and climate change through diverse strategies. (Summary by Maneesh Lingwan @LingwanManeesh) Nature 10.1038/s41586-024-07731-3
Earth’s terrestrial ecosystems harbor remarkable plant diversity. Some of this diversity can be quantified as functional traits, which provide a measure for diversity in different environments. In this new work, Gross et al. used a functional trait analysis to examine how plants respond to increasing aridity and animal grazing in global drylands, which are highly vulnerable to drivers of global change. The study analyzed 133,769 traits from 301 perennial plants across 98 sites in 25 countries. The findings break the widely accepted belief that extreme climatic conditions lead to a decline in the diversity of plant traits. On the contrary, it suggests that plants in arid regions use a wide variety of adaptation strategies, and this diversity increases with the degree of aridity. The authors describe this response to aridity as the “plant loneliness syndrome”. Heavily grazed drylands also correspond to greater diversity in traits. The findings emphasize that factors like soil fertility and plant cover contribute to trait diversity in challenging environments. This study underscores the significance of drylands as a global reservoir of plant diversity and shows how plants can adapt to increasing environmental pressures and climate change through diverse strategies. (Summary by Maneesh Lingwan @LingwanManeesh) Nature 10.1038/s41586-024-07731-3
Plant memories: How people build lasting connections with plants
 People often fail to recognize and appreciate the importance of plants, a phenomenon known as Plant Awareness Disparity (PAD), also known as plant blindness. This lack of awareness has a negative impact on botanical knowledge and skill acquisitions, consequently affecting biodiversity conservation efforts. McGinn et al. investigated the relationship between art and plant awareness through an exhibition, “In Memory of Plants,” at the Alternative Kilkenny Arts Festival 2022, using art as a tool to address PAD phenomenon. The study revealed the importance of art as a tool for reflection and emotional engagement to connect people to plants. Visitors at the exhibition were encouraged to share their personal plant-related memories, and plants were seen to link people to significant experiences and relationships. The analysis demonstrated the importance of sensory experiences in relating to plants, giving a deeper understanding of human-plant connections and buttressing art-based research as an effective tool to enhance plant awareness and stimulate collaboration among artists, botanists and science communicators. This study, therefore advocates for art as a vital instrument in addressing biodiversity loss and supporting plant conservation efforts. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plants People Planet 10.1002/ppp3.10555
People often fail to recognize and appreciate the importance of plants, a phenomenon known as Plant Awareness Disparity (PAD), also known as plant blindness. This lack of awareness has a negative impact on botanical knowledge and skill acquisitions, consequently affecting biodiversity conservation efforts. McGinn et al. investigated the relationship between art and plant awareness through an exhibition, “In Memory of Plants,” at the Alternative Kilkenny Arts Festival 2022, using art as a tool to address PAD phenomenon. The study revealed the importance of art as a tool for reflection and emotional engagement to connect people to plants. Visitors at the exhibition were encouraged to share their personal plant-related memories, and plants were seen to link people to significant experiences and relationships. The analysis demonstrated the importance of sensory experiences in relating to plants, giving a deeper understanding of human-plant connections and buttressing art-based research as an effective tool to enhance plant awareness and stimulate collaboration among artists, botanists and science communicators. This study, therefore advocates for art as a vital instrument in addressing biodiversity loss and supporting plant conservation efforts. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plants People Planet 10.1002/ppp3.10555
 Earth’s terrestrial ecosystems harbor remarkable plant diversity. Some of this diversity can be quantified as functional traits, which provide a measure for diversity in different environments. In this new work, Gross et al. used a functional trait analysis to examine how plants respond to increasing aridity and animal grazing in global drylands, which are highly vulnerable to drivers of global change. The study analyzed 133,769 traits from 301 perennial plants across 98 sites in 25 countries. The findings break the widely accepted belief that extreme climatic conditions lead to a decline in the diversity of plant traits. On the contrary, it suggests that plants in arid regions use a wide variety of adaptation strategies, and this diversity increases with the degree of aridity. The authors describe this response to aridity as the “plant loneliness syndrome”. Heavily grazed drylands also correspond to greater diversity in traits. The findings emphasize that factors like soil fertility and plant cover contribute to trait diversity in challenging environments. This study underscores the significance of drylands as a global reservoir of plant diversity and shows how plants can adapt to increasing environmental pressures and climate change through diverse strategies. (Summary by Maneesh Lingwan @LingwanManeesh) Nature 10.1038/s41586-024-07731-3
Earth’s terrestrial ecosystems harbor remarkable plant diversity. Some of this diversity can be quantified as functional traits, which provide a measure for diversity in different environments. In this new work, Gross et al. used a functional trait analysis to examine how plants respond to increasing aridity and animal grazing in global drylands, which are highly vulnerable to drivers of global change. The study analyzed 133,769 traits from 301 perennial plants across 98 sites in 25 countries. The findings break the widely accepted belief that extreme climatic conditions lead to a decline in the diversity of plant traits. On the contrary, it suggests that plants in arid regions use a wide variety of adaptation strategies, and this diversity increases with the degree of aridity. The authors describe this response to aridity as the “plant loneliness syndrome”. Heavily grazed drylands also correspond to greater diversity in traits. The findings emphasize that factors like soil fertility and plant cover contribute to trait diversity in challenging environments. This study underscores the significance of drylands as a global reservoir of plant diversity and shows how plants can adapt to increasing environmental pressures and climate change through diverse strategies. (Summary by Maneesh Lingwan @LingwanManeesh) Nature 10.1038/s41586-024-07731-3