Virtual issue: The chemical language of plant–microbe–microbe associations
 Don’t miss this exciting Virtual Issue from New Phytologist on “plant-microbe-microbe” interactions. That’s not a typo – many of the articles address the signals that coordinate such multi-factorial interactions, as there is a growing recognition that interrelations between microbes influence how they interaction with plants (see also the review below by Mesny et al.). This Virtual issue centers the role of chemical signals, including hormones, metabolites, peptides, and volatile organic compounds (VOCs), in shaping plant-microbe-microbe outcomes. The editorial beautifully summarizes this exciting topic and introduces some of the key questions still being addressed. One of the articles I found particularly intriguing is “Listening to plant’s Esperanto via root exudates.” Esperanto is a constructed international language that was developed to foster communication between people of diverse origins, and I love thinking of plant exudates as a universal language that can be understood by all microbes. I also enjoyed the article exploring how “Microbial terroir” affects the flavor chemistry of mustard seed. Terroir is a term commonly used in winemaking that refers to how the environment in which a plant is grown influences its chemical composition and flavor. The potential applications of a better understanding plant-microbe-microbe interactions are vast, from optimizing or customizing flavor, to harnessing beneficial microbes in order to suppress disease-causing microbes, and beyond. (Summary by Mary Williams @PlantTeaching) New Phytologist. Editorial 10.1111/nph.20124.
Don’t miss this exciting Virtual Issue from New Phytologist on “plant-microbe-microbe” interactions. That’s not a typo – many of the articles address the signals that coordinate such multi-factorial interactions, as there is a growing recognition that interrelations between microbes influence how they interaction with plants (see also the review below by Mesny et al.). This Virtual issue centers the role of chemical signals, including hormones, metabolites, peptides, and volatile organic compounds (VOCs), in shaping plant-microbe-microbe outcomes. The editorial beautifully summarizes this exciting topic and introduces some of the key questions still being addressed. One of the articles I found particularly intriguing is “Listening to plant’s Esperanto via root exudates.” Esperanto is a constructed international language that was developed to foster communication between people of diverse origins, and I love thinking of plant exudates as a universal language that can be understood by all microbes. I also enjoyed the article exploring how “Microbial terroir” affects the flavor chemistry of mustard seed. Terroir is a term commonly used in winemaking that refers to how the environment in which a plant is grown influences its chemical composition and flavor. The potential applications of a better understanding plant-microbe-microbe interactions are vast, from optimizing or customizing flavor, to harnessing beneficial microbes in order to suppress disease-causing microbes, and beyond. (Summary by Mary Williams @PlantTeaching) New Phytologist. Editorial 10.1111/nph.20124.
Review. Microbial tug-of-war: How plants and pathogens manipulate microbiomes
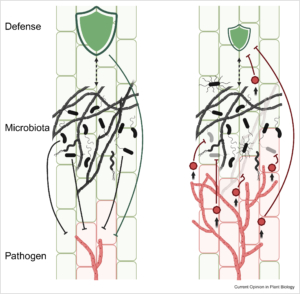 The composition of plant-associated microbes is influenced by plant genetics, immune responses, environmental factors, and interactions between microbes. During disease development, the microbial community at infection sites changes due to tissue damage, altered immune responses, and manipulation via pathogen effector proteins. Mesny et al. argue that host-pathogen co-evolution involves not just the direct interaction between the two, but also the dynamics within the host’s microbiota. For example, effectors produced by fungal pathogens can have antimicrobial properties that suppress protective microbes, enhancing fungal pathogenicity. On the other hand, foliar infections by fungal pathogens like Botrytis cinerea have been shown to stimulate beneficial microbes like Trichoderma harzianum at plant roots. This recruitment of beneficial microbes, termed the “cry-for-help” response, can have long-lasting effects on the soil microbiota, potentially leading to disease-suppressive soils that protect future crops. However, disease suppressiveness takes years to build up, possibly due to antimicrobial effector proteins secreted by pathogens, which slow down the recruitment of beneficial microbes. Future research should focus on understanding these mechanisms and their long- and short-term effects, leading to new approaches for sustainable disease management. (Summary by Kumanan N Govaichelvan, @NGKumanan) Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102622
The composition of plant-associated microbes is influenced by plant genetics, immune responses, environmental factors, and interactions between microbes. During disease development, the microbial community at infection sites changes due to tissue damage, altered immune responses, and manipulation via pathogen effector proteins. Mesny et al. argue that host-pathogen co-evolution involves not just the direct interaction between the two, but also the dynamics within the host’s microbiota. For example, effectors produced by fungal pathogens can have antimicrobial properties that suppress protective microbes, enhancing fungal pathogenicity. On the other hand, foliar infections by fungal pathogens like Botrytis cinerea have been shown to stimulate beneficial microbes like Trichoderma harzianum at plant roots. This recruitment of beneficial microbes, termed the “cry-for-help” response, can have long-lasting effects on the soil microbiota, potentially leading to disease-suppressive soils that protect future crops. However, disease suppressiveness takes years to build up, possibly due to antimicrobial effector proteins secreted by pathogens, which slow down the recruitment of beneficial microbes. Future research should focus on understanding these mechanisms and their long- and short-term effects, leading to new approaches for sustainable disease management. (Summary by Kumanan N Govaichelvan, @NGKumanan) Curr. Opin. Plant Biol. 10.1016/j.pbi.2024.102622
PIN auxin transporters also transport auxin-like herbicides (and more?)
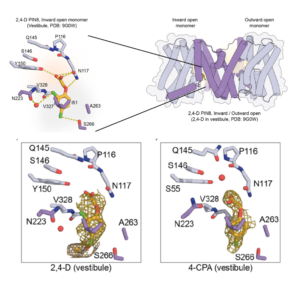 Auxin is an extremely important plant hormone that must be precisely controlled. Auxin-like herbicides have been developed that are often more stable than the natural auxins, thus their application can damage the plant. Here, Schulz, Ung et al. investigated how these auxin herbicides move in plant tissues. The authors investigated the interactions of the well-known PIN auxin transporters and various auxin herbicides, using several methods. Solid supported membrane (SSM) electrophysiology using liposomes as well as expression in oocytes allowed them to measure transport rate and binding affinities of the herbicides. They corroborated these results using cryo-EM, which allowed them to image direct interactions between the herbicides and the transporter proteins. They next generated a set of single residue variants and examined how these affected transport of the herbicides. Their results show that PIN proteins transport auxin herbicides in vivo and in vitro, shed light into the mechanisms of action of the PIN transporters, and open the door to the development of more-efficient herbicides and / or herbicide resistant plants. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.29.610046. (Note that recently a human G-protein-coupled receptor was shown to have structural similarities to plant PIN transporters – maybe like auxin, PINs are everywhere 10.1038/s41586-024-08012-9).
Auxin is an extremely important plant hormone that must be precisely controlled. Auxin-like herbicides have been developed that are often more stable than the natural auxins, thus their application can damage the plant. Here, Schulz, Ung et al. investigated how these auxin herbicides move in plant tissues. The authors investigated the interactions of the well-known PIN auxin transporters and various auxin herbicides, using several methods. Solid supported membrane (SSM) electrophysiology using liposomes as well as expression in oocytes allowed them to measure transport rate and binding affinities of the herbicides. They corroborated these results using cryo-EM, which allowed them to image direct interactions between the herbicides and the transporter proteins. They next generated a set of single residue variants and examined how these affected transport of the herbicides. Their results show that PIN proteins transport auxin herbicides in vivo and in vitro, shed light into the mechanisms of action of the PIN transporters, and open the door to the development of more-efficient herbicides and / or herbicide resistant plants. (Summary by Mary Williams @PlantTeaching) bioRxiv https://doi.org/10.1101/2024.08.29.610046. (Note that recently a human G-protein-coupled receptor was shown to have structural similarities to plant PIN transporters – maybe like auxin, PINs are everywhere 10.1038/s41586-024-08012-9).
Unveiling vacuole biogenesis: Tubular networks are present in plant meristem cells
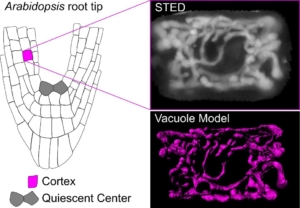 A recent paper by Scheuring and colleagues investigates vacuole biogenesis in meristematic cells of Arabidopsis thaliana, challenging earlier models of vacuole formation. Vacuoles are crucial organelles responsible for various cellular functions, yet their formation has remained puzzling for quite some time. Historically, models have proposed that vacuoles are formed either by contributions from the endoplasmic reticulum (ER) or through homotypic fusion of multivesicular bodies (MVBs). However, recent evidence suggests a more complex process involving multiple membrane sources. The authors use advanced imaging techniques, such as super-resolution stimulated emission depletion (STED) microscopy and fluorescence recovery after photobleaching (FRAP), to confirm the presence of a tubular vacuolar network in meristematic plant cells. The authors introduced a RUBY vacuole reporter line (using betalain fluorescence) to image deeper into tissues, where conventional dyes might not penetrate. This tubular network, observed in young cells near the quiescent center of the root, appears interconnected, which contradicts the theory that MVBs alone are sufficient to form vacuoles. Their findings suggest instead that vacuolar membranes originate from various cellular sources, including the ER. The study further introduces a customized FRAP assay, termed vacuole-connectivity FRAP (vaccFRAP), to analyze vacuolar dynamics. This assay beautifully highlights different connectivity levels within the vacuolar network, providing insights into how vacuoles function as interconnected compartments even in early developmental stages. This research advances our understanding of vacuole formation and its role in plant cell development. (Summary by Ann-Kathrin Rößling @AK_Roessling) Plant Cell 10.1093/plcell/koae243
A recent paper by Scheuring and colleagues investigates vacuole biogenesis in meristematic cells of Arabidopsis thaliana, challenging earlier models of vacuole formation. Vacuoles are crucial organelles responsible for various cellular functions, yet their formation has remained puzzling for quite some time. Historically, models have proposed that vacuoles are formed either by contributions from the endoplasmic reticulum (ER) or through homotypic fusion of multivesicular bodies (MVBs). However, recent evidence suggests a more complex process involving multiple membrane sources. The authors use advanced imaging techniques, such as super-resolution stimulated emission depletion (STED) microscopy and fluorescence recovery after photobleaching (FRAP), to confirm the presence of a tubular vacuolar network in meristematic plant cells. The authors introduced a RUBY vacuole reporter line (using betalain fluorescence) to image deeper into tissues, where conventional dyes might not penetrate. This tubular network, observed in young cells near the quiescent center of the root, appears interconnected, which contradicts the theory that MVBs alone are sufficient to form vacuoles. Their findings suggest instead that vacuolar membranes originate from various cellular sources, including the ER. The study further introduces a customized FRAP assay, termed vacuole-connectivity FRAP (vaccFRAP), to analyze vacuolar dynamics. This assay beautifully highlights different connectivity levels within the vacuolar network, providing insights into how vacuoles function as interconnected compartments even in early developmental stages. This research advances our understanding of vacuole formation and its role in plant cell development. (Summary by Ann-Kathrin Rößling @AK_Roessling) Plant Cell 10.1093/plcell/koae243
Decoding the signaling precision of receptor-MAPK pathways
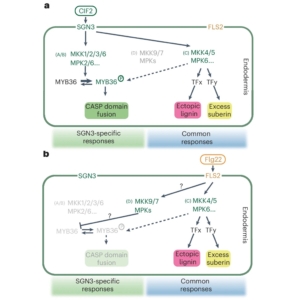 Have you ever wondered how cells distinguish between the diverse array of external signals traveling through similar pathways? To understand the intricacies of plant signaling mechanisms, Ma et al. used single endodermal cells of Arabidopsis roots as a model to compare two receptor pathways. One pathway uses the immune receptor FLAGELLIN SENSING2 (FLS2) to detect microbial signals, while the other involves the SCHENGEN3 (SGN3) receptor, a key regulator of Casparian strip (CS) development in root endodermal cells. The authors used a combination of genetic, molecular, and biochemical approaches to demonstrate that the SGN3 pathway and the FLS2 pathway activate distinct groups of mitogen-activated protein kinase kinases (MKKs). The study further showed that SGN3 stimulation enables CS domain fusion in endodermal cells through MYB36-mediated transcriptional responses, while MYB36 is not activated by the FLS2 pathway. Instead MKK4/5 is activated by FLS2 to facilitate enhanced lignin and suberin deposition. Moreover, FLS2 activates MKK7/9 which are negative regulators of CS development, thus suppressing SGN3-specific responses when this pathway is triggered. Thus, this study sheds light on the intricate complexities of plant signaling mechanisms through single cell analyses. (Summary by Abira Sahu @AbiraSahu) Nature Plants 10.1038/s41477-024-01768-y
Have you ever wondered how cells distinguish between the diverse array of external signals traveling through similar pathways? To understand the intricacies of plant signaling mechanisms, Ma et al. used single endodermal cells of Arabidopsis roots as a model to compare two receptor pathways. One pathway uses the immune receptor FLAGELLIN SENSING2 (FLS2) to detect microbial signals, while the other involves the SCHENGEN3 (SGN3) receptor, a key regulator of Casparian strip (CS) development in root endodermal cells. The authors used a combination of genetic, molecular, and biochemical approaches to demonstrate that the SGN3 pathway and the FLS2 pathway activate distinct groups of mitogen-activated protein kinase kinases (MKKs). The study further showed that SGN3 stimulation enables CS domain fusion in endodermal cells through MYB36-mediated transcriptional responses, while MYB36 is not activated by the FLS2 pathway. Instead MKK4/5 is activated by FLS2 to facilitate enhanced lignin and suberin deposition. Moreover, FLS2 activates MKK7/9 which are negative regulators of CS development, thus suppressing SGN3-specific responses when this pathway is triggered. Thus, this study sheds light on the intricate complexities of plant signaling mechanisms through single cell analyses. (Summary by Abira Sahu @AbiraSahu) Nature Plants 10.1038/s41477-024-01768-y
Tomato SAURs regulate root elongation under Al stress
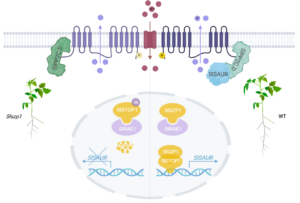 In acidic soils, aluminum (Al) toxicity is a major problem that causes a dramatic arrest in root elongation. Aluminum tolerance can include the secretion of organic acids and the sequestration of Al in internal cellular compartments. Here, Dong et. al shed light on a new pathway that alleviates the inhibition of cell elongation in the presence of Al. Previous studies have highlighted the importance of the plasma-membrane-localized H+-ATPase in Al tolerance. For example, in cucumber, Al decreases the activity of the PM H+-ATPase, whereas in tomato its activity is increased in the presence of Al. In rice, inhibition of the PM H+-ATPase lowers the Al uptake through specific channels, and in Arabidopsis increased PM H+-ATPase activity increases the secretion of organic acids. Previously, a rice mutant was identified, sal1 (sensitive to aluminum 1) that is hypersensitive to aluminum. SAL1 encodes a plasma-membrane localized PP2C.D-type protein phosphatase that interacts with and suppresses the activity of the plasma-membrane-localized H+-ATPase. Previously also, STOP1 was shown to be a transcription factor that is upregulated in the presence of Al and enhances Al tolerance. Here, Dong et al. identified two genes that are transcriptional targets of tomato STOP1 (SlSTOP1) that encode small-auxin upregulated proteins, SlSAUR36 and SlSAUR38. These proteins interact with protein phosphatase SlPP2C.D, lowering its inhibition of PM H+-ATPases and promoting cell expansion in the presence of Al. These insights could contribute to the development of enhanced tolerance to aluminum in tomato and other crops. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae519
In acidic soils, aluminum (Al) toxicity is a major problem that causes a dramatic arrest in root elongation. Aluminum tolerance can include the secretion of organic acids and the sequestration of Al in internal cellular compartments. Here, Dong et. al shed light on a new pathway that alleviates the inhibition of cell elongation in the presence of Al. Previous studies have highlighted the importance of the plasma-membrane-localized H+-ATPase in Al tolerance. For example, in cucumber, Al decreases the activity of the PM H+-ATPase, whereas in tomato its activity is increased in the presence of Al. In rice, inhibition of the PM H+-ATPase lowers the Al uptake through specific channels, and in Arabidopsis increased PM H+-ATPase activity increases the secretion of organic acids. Previously, a rice mutant was identified, sal1 (sensitive to aluminum 1) that is hypersensitive to aluminum. SAL1 encodes a plasma-membrane localized PP2C.D-type protein phosphatase that interacts with and suppresses the activity of the plasma-membrane-localized H+-ATPase. Previously also, STOP1 was shown to be a transcription factor that is upregulated in the presence of Al and enhances Al tolerance. Here, Dong et al. identified two genes that are transcriptional targets of tomato STOP1 (SlSTOP1) that encode small-auxin upregulated proteins, SlSAUR36 and SlSAUR38. These proteins interact with protein phosphatase SlPP2C.D, lowering its inhibition of PM H+-ATPases and promoting cell expansion in the presence of Al. These insights could contribute to the development of enhanced tolerance to aluminum in tomato and other crops. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae519
 In acidic soils, aluminum (Al) toxicity is a major problem that causes a dramatic arrest in root elongation. Aluminum tolerance can include the secretion of organic acids and the sequestration of Al in internal cellular compartments. Here, Dong et. al shed light on a new pathway that alleviates the inhibition of cell elongation in the presence of Al. Previous studies have highlighted the importance of the plasma-membrane-localized H+-ATPase in Al tolerance. For example, in cucumber, Al decreases the activity of the PM H+-ATPase, whereas in tomato its activity is increased in the presence of Al. In rice, inhibition of the PM H+-ATPase lowers the Al uptake through specific channels, and in Arabidopsis increased PM H+-ATPase activity increases the secretion of organic acids. Previously, a rice mutant was identified, sal1 (sensitive to aluminum 1) that is hypersensitive to aluminum. SAL1 encodes a plasma-membrane localized PP2C.D-type protein phosphatase that interacts with and suppresses the activity of the plasma-membrane-localized H+-ATPase. Previously also, STOP1 was shown to be a transcription factor that is upregulated in the presence of Al and enhances Al tolerance. Here, Dong et al. identified two genes that are transcriptional targets of tomato STOP1 (SlSTOP1) that encode small-auxin upregulated proteins, SlSAUR36 and SlSAUR38. These proteins interact with protein phosphatase SlPP2C.D, lowering its inhibition of PM H+-ATPases and promoting cell expansion in the presence of Al. These insights could contribute to the development of enhanced tolerance to aluminum in tomato and other crops. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae519
In acidic soils, aluminum (Al) toxicity is a major problem that causes a dramatic arrest in root elongation. Aluminum tolerance can include the secretion of organic acids and the sequestration of Al in internal cellular compartments. Here, Dong et. al shed light on a new pathway that alleviates the inhibition of cell elongation in the presence of Al. Previous studies have highlighted the importance of the plasma-membrane-localized H+-ATPase in Al tolerance. For example, in cucumber, Al decreases the activity of the PM H+-ATPase, whereas in tomato its activity is increased in the presence of Al. In rice, inhibition of the PM H+-ATPase lowers the Al uptake through specific channels, and in Arabidopsis increased PM H+-ATPase activity increases the secretion of organic acids. Previously, a rice mutant was identified, sal1 (sensitive to aluminum 1) that is hypersensitive to aluminum. SAL1 encodes a plasma-membrane localized PP2C.D-type protein phosphatase that interacts with and suppresses the activity of the plasma-membrane-localized H+-ATPase. Previously also, STOP1 was shown to be a transcription factor that is upregulated in the presence of Al and enhances Al tolerance. Here, Dong et al. identified two genes that are transcriptional targets of tomato STOP1 (SlSTOP1) that encode small-auxin upregulated proteins, SlSAUR36 and SlSAUR38. These proteins interact with protein phosphatase SlPP2C.D, lowering its inhibition of PM H+-ATPases and promoting cell expansion in the presence of Al. These insights could contribute to the development of enhanced tolerance to aluminum in tomato and other crops. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1093/plphys/kiae519