Review: Bringing purple tomatoes to market
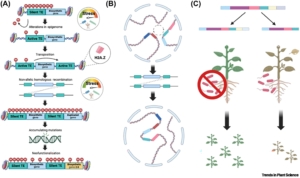
 This interesting article by Cathie Martin and Eugenio Butelli describes the process through which purple, anthocyanin-accumulating tomatoes were brought to the US market, from idea to business plan to regulatory status review and finally direct-to-consumer sales. Although these tomatoes have demonstrated health benefits, the developers had difficulty finding partners who wanted to help commercialize them, so they themselves worked through the long process by first forming a company called Norfolk Plant Sciences (NPS). The article describes the science behind elevated accumulation of anthocyanin, caused by increased expression of two key transcription factors in the metabolic pathway, as well as the characterization of the nutritional qualities of the tomatoes. The remainder of the article is particularly interesting and useful to plant scientists, as it lays out the steps that the authors undertook to get regulatory approval from the USDA/APHIS (Animal and Plant Health Inspection Service). It’s not often we get a first-hand perspective into how a genetically-engineered food can be made available to consumers, so this story will be particularly useful to other small-scale entrepreneurs. (Summary by Mary Williams @plantteaching.bsky.social @PlantTeaching) ACS Food Sci. Technol. 10.1021/acsfoodscitech.4c00692
This interesting article by Cathie Martin and Eugenio Butelli describes the process through which purple, anthocyanin-accumulating tomatoes were brought to the US market, from idea to business plan to regulatory status review and finally direct-to-consumer sales. Although these tomatoes have demonstrated health benefits, the developers had difficulty finding partners who wanted to help commercialize them, so they themselves worked through the long process by first forming a company called Norfolk Plant Sciences (NPS). The article describes the science behind elevated accumulation of anthocyanin, caused by increased expression of two key transcription factors in the metabolic pathway, as well as the characterization of the nutritional qualities of the tomatoes. The remainder of the article is particularly interesting and useful to plant scientists, as it lays out the steps that the authors undertook to get regulatory approval from the USDA/APHIS (Animal and Plant Health Inspection Service). It’s not often we get a first-hand perspective into how a genetically-engineered food can be made available to consumers, so this story will be particularly useful to other small-scale entrepreneurs. (Summary by Mary Williams @plantteaching.bsky.social @PlantTeaching) ACS Food Sci. Technol. 10.1021/acsfoodscitech.4c00692
Metabolites through the looking glass with CEST MRI
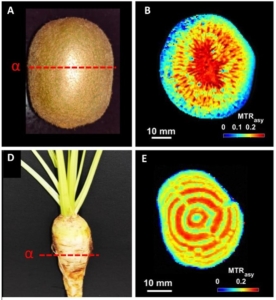 Non-invasive imaging technologies like computed tomography and magnetic resonance imaging (MRI) have revolutionized medicine by improving diagnostics and guiding treatment. Due to its versatility, MRI also holds potential for plant sciences, where it can be used to visualize and quantify metabolites within organs, tissues, and cells. However, challenges specific to plant tissues have hindered its implementation. Mayer et al. addressed these challenges by integrating chemical exchange saturation transfer (CEST), an MRI contrast technique that enhances signal detection and reduces susceptibility to magnetic field disturbances in plant tissues. CEST MRI enables high-resolution visualization of metabolites like amino acids and sugars. The researchers tested their method on various plant systems, from model crops like potato tubers to more complex tissues such as maize kernels. Compared to standard MRI, CEST MRI offers a higher signal-to-noise ratio, better spatial resolution, and faster detection times. Additionally, its ability to image live and complex plant tissues, such as barley grains on the spike, make it a powerful tool for the in vivo detection of metabolites in economically important sink organs including tubers, roots, and grains. Unlike most metabolomics techniques, which measure specific metabolites destructively, CEST MRI captures broad metabolite classes non-invasively, in live tissues, and with high resolution. This remarkable advancement in the field of plant metabolism has exciting potential for plant phenotyping platforms, breeding programs, and biofortification strategies by selecting traits linked to optimal metabolite concentrations. (Summary by Thomas Depaepe @thdpaepe.bsky.social @thdpaepe) Science Advances 10.1126/sciadv.adq4424
Non-invasive imaging technologies like computed tomography and magnetic resonance imaging (MRI) have revolutionized medicine by improving diagnostics and guiding treatment. Due to its versatility, MRI also holds potential for plant sciences, where it can be used to visualize and quantify metabolites within organs, tissues, and cells. However, challenges specific to plant tissues have hindered its implementation. Mayer et al. addressed these challenges by integrating chemical exchange saturation transfer (CEST), an MRI contrast technique that enhances signal detection and reduces susceptibility to magnetic field disturbances in plant tissues. CEST MRI enables high-resolution visualization of metabolites like amino acids and sugars. The researchers tested their method on various plant systems, from model crops like potato tubers to more complex tissues such as maize kernels. Compared to standard MRI, CEST MRI offers a higher signal-to-noise ratio, better spatial resolution, and faster detection times. Additionally, its ability to image live and complex plant tissues, such as barley grains on the spike, make it a powerful tool for the in vivo detection of metabolites in economically important sink organs including tubers, roots, and grains. Unlike most metabolomics techniques, which measure specific metabolites destructively, CEST MRI captures broad metabolite classes non-invasively, in live tissues, and with high resolution. This remarkable advancement in the field of plant metabolism has exciting potential for plant phenotyping platforms, breeding programs, and biofortification strategies by selecting traits linked to optimal metabolite concentrations. (Summary by Thomas Depaepe @thdpaepe.bsky.social @thdpaepe) Science Advances 10.1126/sciadv.adq4424
Crowd control by DCP5 – a new cytoplasmic osmosensor
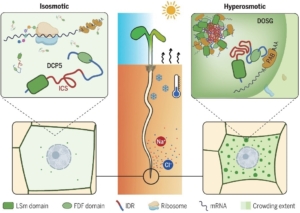 Osmosis, driving water uptake and transport, is crucial for plants. It supports nutrient uptake, turgidity, and overall plant health. In hyperosmotic conditions, caused by drought, salinity, and cold stress, water loss triggers osmotic responses. A key question is: what sensors detect osmotic changes? While traditional sensors are ligand-receptor based or stretch-activated, internal cytoplasmic sensors are also presumed to exist. Recent studies suggest that proteins with intrinsically disordered regions (IDRs) are sensitive to changes in their physicochemical environment. These proteins could therefore serve as sensors that transmit information about the cell’s state. In a recent study, Wang and co-authors identified Arabidopsis Decapping 5 (DCP5) as an intracellular cytoplasmic osmosensor. Their work demonstrates that DCP5 accumulates in cytoplasmic condensates under osmotic or salinity stress, aggregating in a dynamic, reversible manner. They further showed that molecular crowding resulting from volumetric changes – and not other osmotic signals – drives DCP5 aggregation. Additional bioinformatic and genetic analyses revealed that DCP5’s leucine-rich IDR is required for phase separation, driving aggregation and forming stress granules that sequester mRNA molecules. The authors show that these stress granules result in transcriptomic and translational changes that facilitate stress adaptation. In conclusion, Wang and colleagues uncovered a novel mechanism by which plants sense osmotic changes. Their study further highlights important roles for IDRs and protein phase separation in plant stress responses – an area that really deserves more attention. (Summary by Thomas Depaepe @thdpaepe.bsky.social @thdpaepe) Science 10.1126/science.adk9067
Osmosis, driving water uptake and transport, is crucial for plants. It supports nutrient uptake, turgidity, and overall plant health. In hyperosmotic conditions, caused by drought, salinity, and cold stress, water loss triggers osmotic responses. A key question is: what sensors detect osmotic changes? While traditional sensors are ligand-receptor based or stretch-activated, internal cytoplasmic sensors are also presumed to exist. Recent studies suggest that proteins with intrinsically disordered regions (IDRs) are sensitive to changes in their physicochemical environment. These proteins could therefore serve as sensors that transmit information about the cell’s state. In a recent study, Wang and co-authors identified Arabidopsis Decapping 5 (DCP5) as an intracellular cytoplasmic osmosensor. Their work demonstrates that DCP5 accumulates in cytoplasmic condensates under osmotic or salinity stress, aggregating in a dynamic, reversible manner. They further showed that molecular crowding resulting from volumetric changes – and not other osmotic signals – drives DCP5 aggregation. Additional bioinformatic and genetic analyses revealed that DCP5’s leucine-rich IDR is required for phase separation, driving aggregation and forming stress granules that sequester mRNA molecules. The authors show that these stress granules result in transcriptomic and translational changes that facilitate stress adaptation. In conclusion, Wang and colleagues uncovered a novel mechanism by which plants sense osmotic changes. Their study further highlights important roles for IDRs and protein phase separation in plant stress responses – an area that really deserves more attention. (Summary by Thomas Depaepe @thdpaepe.bsky.social @thdpaepe) Science 10.1126/science.adk9067
Many plant transcription factor families have evolutionarily conserved binding motifs
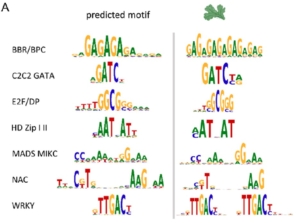 The regulated expression of genes is fundamental to all biological processes, including development, cell growth, and responses to environmental signals. Transcription factors (TFs) are sequence-specific DNA-binding proteins that play a central role in transcriptional regulation by directly interacting with cis-regulatory elements of gene targets. During evolution, the specificity of these interactions can shift due to genome variations or gene duplications. Such changes have been particularly pronounced in yeast, whereas they appear more conserved in flies and humans. To explore this in plants, Zenker and colleagues revisited existing plant TF databases and recalculated TF binding consensus sequences using a standardized pipeline. By comparing angiosperm species with the bryophyte Marchantia polymorpha, their analyses revealed a spectrum of sequence conservation across different TF families, with some binding motifs showing remarkable conservation, tracing back 450 million years. While the relative contributions of cis– and trans-regulation might be challenging to disentangle, this comprehensive catalog of plant TF binding motifs offers valuable insights for studying neofunctionalization and improving TF annotation in emerging plant models. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2024.10.31.621407
The regulated expression of genes is fundamental to all biological processes, including development, cell growth, and responses to environmental signals. Transcription factors (TFs) are sequence-specific DNA-binding proteins that play a central role in transcriptional regulation by directly interacting with cis-regulatory elements of gene targets. During evolution, the specificity of these interactions can shift due to genome variations or gene duplications. Such changes have been particularly pronounced in yeast, whereas they appear more conserved in flies and humans. To explore this in plants, Zenker and colleagues revisited existing plant TF databases and recalculated TF binding consensus sequences using a standardized pipeline. By comparing angiosperm species with the bryophyte Marchantia polymorpha, their analyses revealed a spectrum of sequence conservation across different TF families, with some binding motifs showing remarkable conservation, tracing back 450 million years. While the relative contributions of cis– and trans-regulation might be challenging to disentangle, this comprehensive catalog of plant TF binding motifs offers valuable insights for studying neofunctionalization and improving TF annotation in emerging plant models. (Summary by Ching Chan @ntnuchanlab) bioRxiv 10.1101/2024.10.31.621407
Changes in regulatory regions shape C3 to C4 evolution
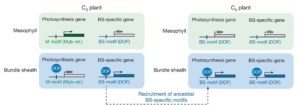 In most land plants, carbon fixation into a three-carbon compound by the enzyme Rubisco takes place in the leaf mesophyll cells; these are called C3 plants. However, a different and more efficient pathway has evolved independently many times, in which a four-carbon metabolite is first produced in the mesophyll cells, followed by Rubisco activity in the bundle sheath cells surrounding the leaf veins; such plants are called C4 plants. While mesophyll and bundle sheath cells exist in both C3 and C4 plants, the specialization of bundle sheath cells for photosynthesis is a particular feature of C4 species. Swift, Luginbuehl et al. focused on rice (C3) and sorghum (C4) to elucidate the molecular changes underlying the specialization of bundle sheath cells for photosynthetic functions. The authors found that genes showing specialized expression in sorghum bundle sheath cells show an enrichment in DOF (DNA binding with one finger) binding motifs. Such motifs are absent in the regulatory regions of their mesophyll-specific rice counterparts, despite a similar expression of DOF transcription factors in the two species. Based on their findings, the authors propose a general model whereby changes in the promoter of photosynthesis genes of C4 plants, and in particular an enrichment in DOF motifs, control expression of such genes in bundle sheath cells instead of mesophyll cells. In C3 plants, different motifs in the promoter of photosynthesis genes drive their expression in mesophyll cells but not in bundle sheath cells. This work provides new insights into the transcriptional changes associated with C4 photosynthesis at the single-cell level, and will contribute to international efforts to engineer C4 photosynthesis in C3 plants to increase their efficiency. Moreover, it provides an example of the importance of changes in cis-regulatory sequences to shape important evolutionary traits. Summary by Laura Turchi, @turchil.bsky.social @turchi_l) Nature 10.1038/s41586-024-08204-3
In most land plants, carbon fixation into a three-carbon compound by the enzyme Rubisco takes place in the leaf mesophyll cells; these are called C3 plants. However, a different and more efficient pathway has evolved independently many times, in which a four-carbon metabolite is first produced in the mesophyll cells, followed by Rubisco activity in the bundle sheath cells surrounding the leaf veins; such plants are called C4 plants. While mesophyll and bundle sheath cells exist in both C3 and C4 plants, the specialization of bundle sheath cells for photosynthesis is a particular feature of C4 species. Swift, Luginbuehl et al. focused on rice (C3) and sorghum (C4) to elucidate the molecular changes underlying the specialization of bundle sheath cells for photosynthetic functions. The authors found that genes showing specialized expression in sorghum bundle sheath cells show an enrichment in DOF (DNA binding with one finger) binding motifs. Such motifs are absent in the regulatory regions of their mesophyll-specific rice counterparts, despite a similar expression of DOF transcription factors in the two species. Based on their findings, the authors propose a general model whereby changes in the promoter of photosynthesis genes of C4 plants, and in particular an enrichment in DOF motifs, control expression of such genes in bundle sheath cells instead of mesophyll cells. In C3 plants, different motifs in the promoter of photosynthesis genes drive their expression in mesophyll cells but not in bundle sheath cells. This work provides new insights into the transcriptional changes associated with C4 photosynthesis at the single-cell level, and will contribute to international efforts to engineer C4 photosynthesis in C3 plants to increase their efficiency. Moreover, it provides an example of the importance of changes in cis-regulatory sequences to shape important evolutionary traits. Summary by Laura Turchi, @turchil.bsky.social @turchi_l) Nature 10.1038/s41586-024-08204-3
Regulatory mechanisms of strigolactone perception in rice
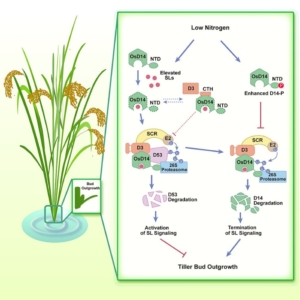 Strigolactones (SLs), a group of carotenoid-derived small signaling molecules and a class of phytohormones, play key roles in regulating various growth and developmental processes in plants. Additionally, they act as rhizosphere signaling molecules that promote symbiosis with arbuscular mycorrhizal (AM) fungi. Despite the identification of key components involved in SL perception and signaling through genetic and biochemical assays, the molecular mechanisms underlying SL signaling remain only partially understood. In a recent study, Hu and colleagues investigated DWARF14 (D14), a crucial SL receptor, and its associated receptor complex components. Their findings reveal that the formation of the D14-D3-ASK1 complex is essential for activating SL perception. SL induces the ubiquitination and degradation of D14, a process that requires direct interaction with D3. This regulatory mechanism is particularly significant in rice under low nitrogen conditions, where it affects tillering. This study provides new insights into the multi-layered regulation of SL signaling and opens opportunities for crop improvement and breeding programs. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2024.10.009
Strigolactones (SLs), a group of carotenoid-derived small signaling molecules and a class of phytohormones, play key roles in regulating various growth and developmental processes in plants. Additionally, they act as rhizosphere signaling molecules that promote symbiosis with arbuscular mycorrhizal (AM) fungi. Despite the identification of key components involved in SL perception and signaling through genetic and biochemical assays, the molecular mechanisms underlying SL signaling remain only partially understood. In a recent study, Hu and colleagues investigated DWARF14 (D14), a crucial SL receptor, and its associated receptor complex components. Their findings reveal that the formation of the D14-D3-ASK1 complex is essential for activating SL perception. SL induces the ubiquitination and degradation of D14, a process that requires direct interaction with D3. This regulatory mechanism is particularly significant in rice under low nitrogen conditions, where it affects tillering. This study provides new insights into the multi-layered regulation of SL signaling and opens opportunities for crop improvement and breeding programs. (Summary by Ching Chan @ntnuchanlab) Cell 10.1016/j.cell.2024.10.009
Epigenetic clocks in plants: uncovering DNA methylation decay in aging
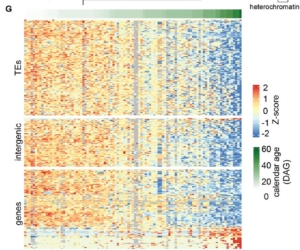 What if plants could teach us about aging? Understanding how and why living organisms age is a fundamental question in biology and medicine. While most research focuses on humans, the model plant Arabidopsis thaliana offers unique insights into how organisms age. In a recent study, Dai et al. analyzed first true leaves at various ages and observed an intriguing pattern: a progressive loss of methylation integrity in heterochromatin regions as the tissues aged. This loss reflects patterns observed in human aging and diseases like cancer. These cumulative methylation losses result in significant differentially methylated regions in senescent leaves, accompanied by an increase in transposable element transcripts. This finding not only highlights the ability of plants to decouple biological and chronological age but also introduces an exciting parallel to the “epigenetic clocks” used to predict biological age in mammals. Even more fascinating is the discovery that new organs are epigenetically young and can age at independent rates from the rest of the organism, suggesting that meristems are “ageless,” producing germline cells and somatic organs that age independently. The study also identifies key regulatory genes, such as TCX5 and TCX6, which act as molecular switches repressing DNA methylation maintenance during aging. Mutants for this mechanism prevent epigenetic aging entirely, opening doors to understanding—and potentially manipulating—aging at a fundamental level. Lastly, but just as importantly, this study highlights that both the organ’s age and the timing of its emergence are critical factors that must be carefully accounted for when sampling! This work emphasizes the tremendous potential of Arabidopsis as a model species in the field of aging research. (Summary by Ileana Tossolini @IleanaDrt @ileanadrt.bsky.social) bioRxiv https://www.biorxiv.org/content/10.1101/2024.11.04.621941v1
What if plants could teach us about aging? Understanding how and why living organisms age is a fundamental question in biology and medicine. While most research focuses on humans, the model plant Arabidopsis thaliana offers unique insights into how organisms age. In a recent study, Dai et al. analyzed first true leaves at various ages and observed an intriguing pattern: a progressive loss of methylation integrity in heterochromatin regions as the tissues aged. This loss reflects patterns observed in human aging and diseases like cancer. These cumulative methylation losses result in significant differentially methylated regions in senescent leaves, accompanied by an increase in transposable element transcripts. This finding not only highlights the ability of plants to decouple biological and chronological age but also introduces an exciting parallel to the “epigenetic clocks” used to predict biological age in mammals. Even more fascinating is the discovery that new organs are epigenetically young and can age at independent rates from the rest of the organism, suggesting that meristems are “ageless,” producing germline cells and somatic organs that age independently. The study also identifies key regulatory genes, such as TCX5 and TCX6, which act as molecular switches repressing DNA methylation maintenance during aging. Mutants for this mechanism prevent epigenetic aging entirely, opening doors to understanding—and potentially manipulating—aging at a fundamental level. Lastly, but just as importantly, this study highlights that both the organ’s age and the timing of its emergence are critical factors that must be carefully accounted for when sampling! This work emphasizes the tremendous potential of Arabidopsis as a model species in the field of aging research. (Summary by Ileana Tossolini @IleanaDrt @ileanadrt.bsky.social) bioRxiv https://www.biorxiv.org/content/10.1101/2024.11.04.621941v1
Disrupting plant inositol pyrophosphates to mitigate P pollution
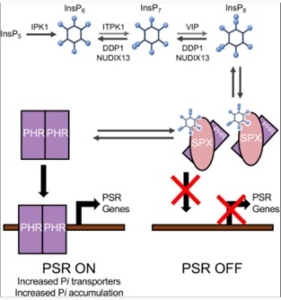 Inositol pyrophosphates (PP-InsP) are plant signaling molecules that regulate phosphate homeostasis and related metabolic processes. But a global phosphate crisis, due to resource depletion and environmental pollution from excess phosphate, has raised the need for phosphate management. Freed et al. investigated strategies for enhancing phosphate management by manipulating PP-InsP signal pathways using both synthetic and native genes in Arabidopsis and pennycress, a cover crop. Their findings revealed that expressing Diadenosine and Diphosphoinositol Polyphosphate Phosphohydrolase (DDP1), a yeast enzyme, decreases PP-InsP levels, activates phosphate starvation response, and enhances phosphate accumulation in Arabidopsis and pennycress. These results suggest that altering plant PP-InsP levels can be a strategy through which plants can be engineered to reclaim phosphate from soils. A native Arabidopsis gene, Nucleoside diphosphate-linked moiety X 13 (NUDIX13), which encodes an enzyme similar to DDP1, was also identified as promising target for regulating PP-InsP pathways. This study suggests the possibility of using DDP1 and NUDIX13 as tools for development of plants to manage phosphate resources and mitigate environmental impacts of excess phosphate, therefore, addressing phosphate pollution issues. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Physiol. 10.1093/plphys/kiae582
Inositol pyrophosphates (PP-InsP) are plant signaling molecules that regulate phosphate homeostasis and related metabolic processes. But a global phosphate crisis, due to resource depletion and environmental pollution from excess phosphate, has raised the need for phosphate management. Freed et al. investigated strategies for enhancing phosphate management by manipulating PP-InsP signal pathways using both synthetic and native genes in Arabidopsis and pennycress, a cover crop. Their findings revealed that expressing Diadenosine and Diphosphoinositol Polyphosphate Phosphohydrolase (DDP1), a yeast enzyme, decreases PP-InsP levels, activates phosphate starvation response, and enhances phosphate accumulation in Arabidopsis and pennycress. These results suggest that altering plant PP-InsP levels can be a strategy through which plants can be engineered to reclaim phosphate from soils. A native Arabidopsis gene, Nucleoside diphosphate-linked moiety X 13 (NUDIX13), which encodes an enzyme similar to DDP1, was also identified as promising target for regulating PP-InsP pathways. This study suggests the possibility of using DDP1 and NUDIX13 as tools for development of plants to manage phosphate resources and mitigate environmental impacts of excess phosphate, therefore, addressing phosphate pollution issues. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Physiol. 10.1093/plphys/kiae582
A trade-off between investment in molecular defense repertoires and growth in plants
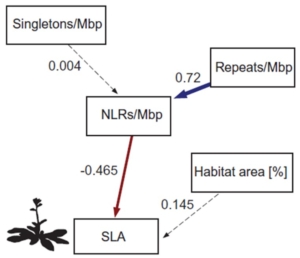 The reduction in crop yields caused by pathogens and pests poses a significant challenge to global food security. Genetic engineering, aimed at strengthening plant defense mechanisms, offers a cost-effective solution for disease control. However, this approach often comes with a growth penalty, commonly referred to as the growth-defense trade-off. Despite extensive research, the precise molecular mechanisms underlying this phenomenon remain incompletely understood. A recent breakthrough by Giolai and Laine highlights the importance of intra- and interspecific variation in observing the growth-defense trade-off. By analyzing the relationship between R-gene density and growth traits, the authors demonstrated that elevated genomic R-gene density is negatively associated with growth in wild plants but not in agricultural or domesticated plants. This finding suggests that maintaining diverse defense repertoires shapes R-gene evolution in wild plant species, balancing growth and defense to maximize fitness and survival. However, this life-history-driven variation appears to be highly sensitive to domestication, underscoring the profound impact of agricultural practices on plant evolutionary dynamics. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.adn2779
The reduction in crop yields caused by pathogens and pests poses a significant challenge to global food security. Genetic engineering, aimed at strengthening plant defense mechanisms, offers a cost-effective solution for disease control. However, this approach often comes with a growth penalty, commonly referred to as the growth-defense trade-off. Despite extensive research, the precise molecular mechanisms underlying this phenomenon remain incompletely understood. A recent breakthrough by Giolai and Laine highlights the importance of intra- and interspecific variation in observing the growth-defense trade-off. By analyzing the relationship between R-gene density and growth traits, the authors demonstrated that elevated genomic R-gene density is negatively associated with growth in wild plants but not in agricultural or domesticated plants. This finding suggests that maintaining diverse defense repertoires shapes R-gene evolution in wild plant species, balancing growth and defense to maximize fitness and survival. However, this life-history-driven variation appears to be highly sensitive to domestication, underscoring the profound impact of agricultural practices on plant evolutionary dynamics. (Summary by Ching Chan @ntnuchanlab) Science 10.1126/science.adn2779
 Inositol pyrophosphates (PP-InsP) are plant signaling molecules that regulate phosphate homeostasis and related metabolic processes. But a global phosphate crisis, due to resource depletion and environmental pollution from excess phosphate, has raised the need for phosphate management. Freed et al. investigated strategies for enhancing phosphate management by manipulating PP-InsP signal pathways using both synthetic and native genes in Arabidopsis and pennycress, a cover crop. Their findings revealed that expressing Diadenosine and Diphosphoinositol Polyphosphate Phosphohydrolase (DDP1), a yeast enzyme, decreases PP-InsP levels, activates phosphate starvation response, and enhances phosphate accumulation in Arabidopsis and pennycress. These results suggest that altering plant PP-InsP levels can be a strategy through which plants can be engineered to reclaim phosphate from soils. A native Arabidopsis gene, Nucleoside diphosphate-linked moiety X 13 (NUDIX13), which encodes an enzyme similar to DDP1, was also identified as promising target for regulating PP-InsP pathways. This study suggests the possibility of using DDP1 and NUDIX13 as tools for development of plants to manage phosphate resources and mitigate environmental impacts of excess phosphate, therefore, addressing phosphate pollution issues. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Physiol. 10.1093/plphys/kiae582
Inositol pyrophosphates (PP-InsP) are plant signaling molecules that regulate phosphate homeostasis and related metabolic processes. But a global phosphate crisis, due to resource depletion and environmental pollution from excess phosphate, has raised the need for phosphate management. Freed et al. investigated strategies for enhancing phosphate management by manipulating PP-InsP signal pathways using both synthetic and native genes in Arabidopsis and pennycress, a cover crop. Their findings revealed that expressing Diadenosine and Diphosphoinositol Polyphosphate Phosphohydrolase (DDP1), a yeast enzyme, decreases PP-InsP levels, activates phosphate starvation response, and enhances phosphate accumulation in Arabidopsis and pennycress. These results suggest that altering plant PP-InsP levels can be a strategy through which plants can be engineered to reclaim phosphate from soils. A native Arabidopsis gene, Nucleoside diphosphate-linked moiety X 13 (NUDIX13), which encodes an enzyme similar to DDP1, was also identified as promising target for regulating PP-InsP pathways. This study suggests the possibility of using DDP1 and NUDIX13 as tools for development of plants to manage phosphate resources and mitigate environmental impacts of excess phosphate, therefore, addressing phosphate pollution issues. (Summary by Idowu Arinola Obisesan, @IdowuAobisesan) Plant Physiol. 10.1093/plphys/kiae582