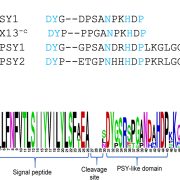
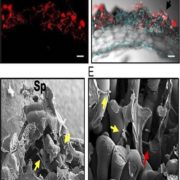
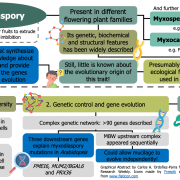
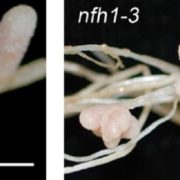
Origin of Plant R Genes
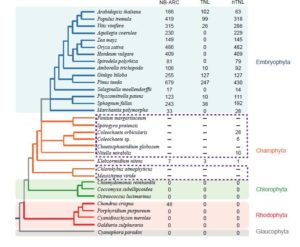 Plants rely on two branches of the innate immunity system to prevent or eliminate microbial infections: one involves cell surface receptors to respond to pathogen- or microbe- associated molecular patterns, and the other acts inside plant cells by using proteins with nucleotide-binding site (NBS) and Leu-rich repeat (LRR) domains. NBS-LRR proteins confer recognition of pathogen effectors either directly or indirectly and trigger disease resistance. Most of the plant disease-resistance genes (R-genes) cloned so far encode NBS-LRR proteins. NBS-LRR proteins are typically classified into two major subfamilies based on the presence or absence of an N-terminal signaling domain. Those possessing the Toll/IL receptor (TIR) domain are referred to as TIR-NBS-LRR proteins (TNLs); those without this domain are referred to as non-TIR-NBS-LRR proteins (nTNLs). Comparative genomic analyses show that R-genes are widely distributed in land plants. However, no R-genes have been reported in algae to date. Thus, plant R-genes are generally thought to have originated in land plants but this idea appears mistaken. Gao et al (10.1104/pp.18.00185) have performed comparative genomic and phylogenetic analyses of R-genes in a wide variety of plants, with an emphasis on basal-branching plants. They report on the presence of of R-genes in the genomes of basal- branching streptophytes, including charophytes, liverworts, and mosses. Phylogenetic analyses suggest that plant R-genes originated in charophytes and R-proteins diversified into TIR-NBS-LRR proteins and non-TIR-NBS-LRR proteins in charophytes. Moreover, R-proteins evolved in a modular fashion through frequent gain or loss of protein domains. Most of the R-genes in basal-branching streptophytes underwent adaptive evolution, indicating an ancient involvement of R-genes in plant-pathogen interactions. These findings provide novel insights into the origin and evolution of R-genes and the mechanisms underlying colonization of terrestrial environments by plants.
Plants rely on two branches of the innate immunity system to prevent or eliminate microbial infections: one involves cell surface receptors to respond to pathogen- or microbe- associated molecular patterns, and the other acts inside plant cells by using proteins with nucleotide-binding site (NBS) and Leu-rich repeat (LRR) domains. NBS-LRR proteins confer recognition of pathogen effectors either directly or indirectly and trigger disease resistance. Most of the plant disease-resistance genes (R-genes) cloned so far encode NBS-LRR proteins. NBS-LRR proteins are typically classified into two major subfamilies based on the presence or absence of an N-terminal signaling domain. Those possessing the Toll/IL receptor (TIR) domain are referred to as TIR-NBS-LRR proteins (TNLs); those without this domain are referred to as non-TIR-NBS-LRR proteins (nTNLs). Comparative genomic analyses show that R-genes are widely distributed in land plants. However, no R-genes have been reported in algae to date. Thus, plant R-genes are generally thought to have originated in land plants but this idea appears mistaken. Gao et al (10.1104/pp.18.00185) have performed comparative genomic and phylogenetic analyses of R-genes in a wide variety of plants, with an emphasis on basal-branching plants. They report on the presence of of R-genes in the genomes of basal- branching streptophytes, including charophytes, liverworts, and mosses. Phylogenetic analyses suggest that plant R-genes originated in charophytes and R-proteins diversified into TIR-NBS-LRR proteins and non-TIR-NBS-LRR proteins in charophytes. Moreover, R-proteins evolved in a modular fashion through frequent gain or loss of protein domains. Most of the R-genes in basal-branching streptophytes underwent adaptive evolution, indicating an ancient involvement of R-genes in plant-pathogen interactions. These findings provide novel insights into the origin and evolution of R-genes and the mechanisms underlying colonization of terrestrial environments by plants.