NPR1 has everything under control
Amna Mhamdi
Ghent University, Department of Plant Biotechnology and Bioinformatics, and VIB Center for Plant Systems Biology, 9052 Ghent, Belgium
Address correspondence to [email protected]
NONEXPRESSER OF PR GENES 1 (NPR1) controls plant immunity and is key to salicylic acid (SA)-dependent signaling pathways. First identified via a genetic screen, NPR1 is named after the npr1 mutants that failed to activate PATHOGENESIS-RELATED (PR) gene expression and were, therefore, defective in resistance to virulent bacteria in local and systemic tissues (Cao et al., 1994; Shah et al., 1997). Although the role of Arabidopsis (Arabidopsis thaliana) NPR1 has been studied for decades it is only relatively recently that NPR1 emerged as a bona fide receptor for SA (Fu et al., 2012; Wu et al., 2012; Ding et al., 2018).
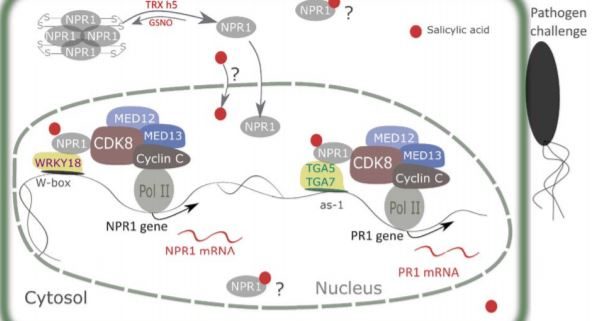
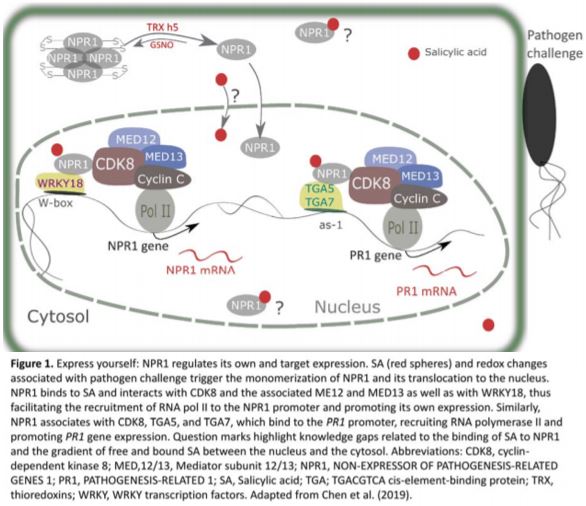 In the absence of SA, NPR1 is cytosolic and exists as oligomers held together by intermolecular disulfide bridges. When plants are challenged with pathogens, SA accumulation and associated redox changes trigger the reduction of NPR1 and its translocation to the nucleus. The reduced NPR1 interacts with transcription factors and enables the expression of target genes (Després et al., 2003; Mou et al., 2003; Tada et al., 2008; Lindermayr et al., 2010). Yet, despite its importance in immunity, there remain several gaps in our understanding of how NPR1 expression is regulated. In this issue of Plant Physiology, Chen and colleagues shed light on this question and reveal a novel mechanism by which NPR1 controls its own expression and that of its target genes (Chen et al., 2019). The authors first demonstrate that a functional NPR1 protein promotes NPR1 gene expression by binding to its promoter. NPR1 interacts with WRKY18 transcription factors and with CYCLIN-DEPENDENT KINASE 8 (CDK8) to regulate its own transcription. In this model, SA promotes an interaction between NPR1 and both partners. In addition, Chen et al. investigated the roles of CDK8 and the associated mediator subunits 12 and 13 in recruiting RNA polymerase II to the promoter of NPR1 and its PR1 target. Analysis of loss-of-function mutants revealed that CDK8 positively regulates NPR1 and PR1 gene expression and is required to fully induce SA responses, although this CDK8 function is independent from its kinase activity (Figure 1).
In the absence of SA, NPR1 is cytosolic and exists as oligomers held together by intermolecular disulfide bridges. When plants are challenged with pathogens, SA accumulation and associated redox changes trigger the reduction of NPR1 and its translocation to the nucleus. The reduced NPR1 interacts with transcription factors and enables the expression of target genes (Després et al., 2003; Mou et al., 2003; Tada et al., 2008; Lindermayr et al., 2010). Yet, despite its importance in immunity, there remain several gaps in our understanding of how NPR1 expression is regulated. In this issue of Plant Physiology, Chen and colleagues shed light on this question and reveal a novel mechanism by which NPR1 controls its own expression and that of its target genes (Chen et al., 2019). The authors first demonstrate that a functional NPR1 protein promotes NPR1 gene expression by binding to its promoter. NPR1 interacts with WRKY18 transcription factors and with CYCLIN-DEPENDENT KINASE 8 (CDK8) to regulate its own transcription. In this model, SA promotes an interaction between NPR1 and both partners. In addition, Chen et al. investigated the roles of CDK8 and the associated mediator subunits 12 and 13 in recruiting RNA polymerase II to the promoter of NPR1 and its PR1 target. Analysis of loss-of-function mutants revealed that CDK8 positively regulates NPR1 and PR1 gene expression and is required to fully induce SA responses, although this CDK8 function is independent from its kinase activity (Figure 1).
Interestingly, Chen et al. (2019) also throw new light onto how NPR1 regulates PR1 expression. NPR1 forms a complex with TGACG cis-element-binding protein (TGA) transcription factors TGA5, TGA7 and CDK8, which forms a bridge between the RNA polymerase II and the PR1 promoter and consequently facilitates PR1 gene transcription. Collectively, these findings enhance our understanding of the mechanisms by which NPR1, a master regulator of immune response, regulates the expression of itself and target genes (Figure 1).
So, do we now fully understand how increased SA availability activate NPR1-dependent and independent gene transcription? Probably only partly. This exciting study raises many questions. How does SA facilitate the binding of NPR1 to the respective interacting proteins? How does NPR1 compete with other potential SA receptors (e.g. NPR3 and NPR4) to promote its own expression? Is direct binding of SA to NPR1 required for controlling its expression and does it occur only at NPR1 or is it necessary for recruiting CDK8 and transcription factors to NPR1? Answers will presumably be found by studying the affinity of NPR1 for SA and the different protein domains or redox sensitive residues that might determine the conformational changes dictated by SA. The future development of in vivo SA detection techniques should also improve our knowledge on how SA gradients between the cytosol and nucleus are maintained and how direct binding of SA to transcription regulators facilitates myriad functions related to hormone-mediated immune responses.
Redox regulation occurs at different nodes to facilitate the establishment of plant immunity (Mou et al., 2003; Després et al., 2003; Lindermayr et al., 2010; Wu et al., 2012), and many post-transcriptional and post-translational modifications shape NPR1-mediated signaling (Spoel et al., 2009; Saleh et al., 2015; Ding et al., 2018). Nevertheless, it remains unclear how these different modifications are integrated to tune the function of the fascinating NPR1. Another intriguing question is to what extent pathogen effectors might target the NPR1 signaling module to hijack plant immunity.