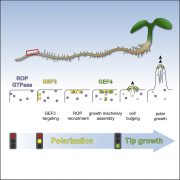
Natural Artist: How a Protein Kinase Helps Sculpt the Pollen Grain Surface From the Inside Out
Finding genes that function in plant development often requires mutant screening, but probing the wealth of natural variation can provide important insights as well. A major focus of developmental biology is uncovering the mechanism behind cell polarity, that is, how components are deposited asymmetrically in a cell at precisely the right place and time. An excellent example of a polar cell is the pollen grain, whose elaborately sculpted exine layer is laid out in beautiful, asymmetric, species-specific patterns. This decay-resistant layer includes gaps (apertures) that serve as sites for pollen tube germination and water transport. Arabidopsis thaliana pollen contains three evenly spaced, groove-like apertures along its surface whose positions are mirrored by the protein INAPERTURATE POLLEN1 (INP1). During pollen development, INP1 assembles in three lines along the plasma membrane (PM) near the overlying callose wall to prevent exine deposition (Dobritsa et al., 2018). However, higher levels of INP1 do not lead to the formation of more or wider apertures, suggesting that it helps execute but does not control aperture formation.
Lee et al. (2018) looked for other players in this process by screening a collection of 316 natural Arabidopsis accessions for abnormally shaped pollen, an indicator of altered pollen aperture. The Libyan accession Martuba (Mt-0) fit the bill, with rounded pollen grains whose apertures are partially covered with exine or sometimes absent. A cross between Mt-0 and the inp1 mutant indicated that Mt-0 harbors a lesion in a gene other than INP1. Positional cloning based on sequence information for Mt-0 revealed a lesion in a 38-gene region. The authors compared non-synonymous polymorphisms between Mt-0 and 62 accessions scored as wild type and narrowed the lesion to a region containing D6 PROTEIN KINASE-LIKE3 (D6PKL3), as confirmed by T-DNA insertion and chemically induced mutant analysis. Complementation of a d6pkl3 mutant with D6PKL3 verified its identity as the gene behind the variation in pollen aperture formation. An examination of other D6PK subfamily mutants showed that only D6PKL3 affects pollen exine morphology.
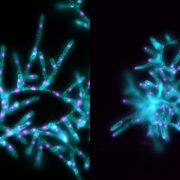
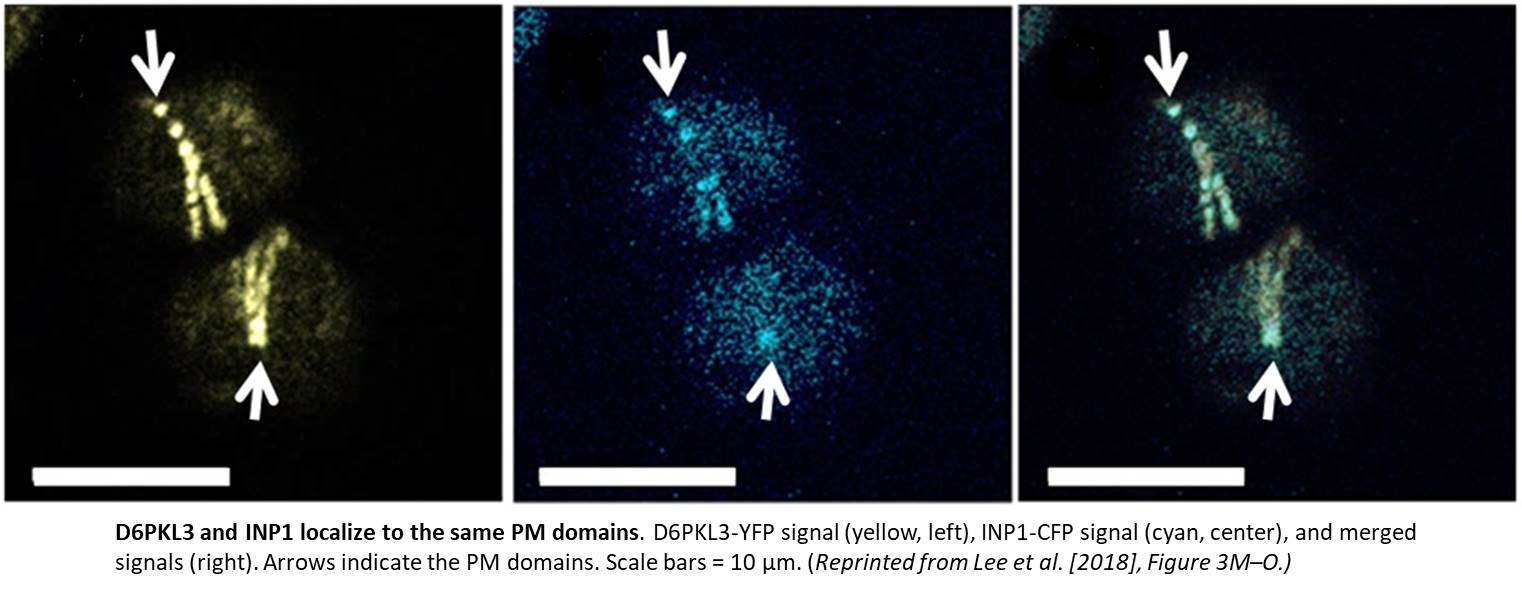
Imagine the excitement when the authors transformed plants with a construct harboring yellow fluorescent protein (YFP) driven by the D6PKL3 promoter and found that it localized to three equally spaced lines in developing pollen grains that overlapped with the location of cyan fluorescent protein (CFP)-labeled INP1 (see figure). D6PKL3 gathered at these regions prior to INP1 accumulation, pointing to its earlier role in aperture formation. In transgenic lines expressing D6PKL3-YFP, this protein often assembled into double parallel lines, and double apertures frequently formed. The unusual aperture phenotypes correlated with increased levels of D6PKL3-YFP expression, suggesting that the dosage of this protein may be a key determinant of polar PM domain and aperture formation during pollen development. The use of a kinase-dead form of D6PKL3 demonstrated that its kinase activity is essential for its correct localization as well as aperture formation.
In d6pkl3 mutants, INP1 failed to aggregate at aperture sites, suggesting that D6PKL3 is required to attract INP1 to its proper location. Conversely, D6PKL3-YFP failed to assemble into peripheral lines when introduced into the inp1 background. However, INP1 and D6PKL3 did not physically interact in various assays. While INP1 is associated with the callose wall, D6PKL3 appears to be associated with the cytoplasmic side of the PM at the aperture domains. How they arrive at these positions is currently unknown, but phosphoinositide lipid species known to be enriched at aperture PM domains could serve as molecular anchors for D6PKL3 and/or INP1.
Which proteins are phosphorylated by D6PKL3 during aperture formation also remains unclear. Discovering whether and how D6PKL3 and INP1 help sculpt the array of intricate pollen exine patterns in other plant species awaits further study.
REFERENCES
Dobritsa, A.A., Kirkpatrick, A.B., Reeder, S.H., Li, P., and Owen, H.A. (2018). Pollen aperture factor INP1 acts late in aperture formation by excluding specific membrane domains from exine deposition. Plant Physiol. 176: 326–339.
Lee, B.H., Weber, Z.T., Zourelidou, M., Hofmeister, B.T., Schmitz, R.J., Schwechheimer, C., and Dobritsa, A.A. (2018). Arabidopsis protein kinase D6PKL3 is involved in the formation of distinct plasma-membrane aperture domains on the pollen surface. Plant Cell 30: doi:10.1015/tpc18.00442.