Molecular switch architecture determines response properties of signaling pathways (PNAS)
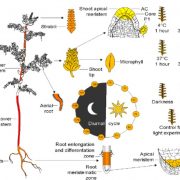
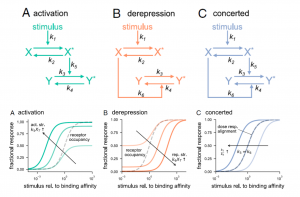 Genetic studies have provided us with countless examples of regulatory switches that transduce a signal into a response. Mutant analysis is usually sufficient to identify these controlling elements, but often in an all-or-nothing way. Here, Ghusinga et al. have taken a theoretical kinetic approach to examine how three types of switch architectures affect the properties of the signaling pathway, using a two-tier model. The first tier is the activation of a receptor by a stimulus, denoted as X -> X*. The second tier is the toggling of the switch between inactive (Y) and active (Y*) state, determined by the kinetics of activation and de-activation. In the activation model, the rate of switch activation is correlated with X*; in other words, active receptor turns the switch on. In the derepression model, the rate of switch deactivation is correlated with X; in other words, inactive receptor turns the switch off. The third model, the concerted model, combines activation and derepression. The authors then examine the sensitivity of each switch type to stimulus amount (e.g., receptor occupancy; dose-response curves), how quickly the response changes with increasing stimulus, and sensitivity to fluctuations. The authors describe the consequences and situational values of each type of switch (as an example, the activation model is sensitive and fast but can lead to false positives, making it a good fit for the flight-or-fight adrenaline response). By contrast, slower derepression signaling is more common in resource-limited plants. This analysis will contribute the development of quantitative models of plants. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2013401118
Genetic studies have provided us with countless examples of regulatory switches that transduce a signal into a response. Mutant analysis is usually sufficient to identify these controlling elements, but often in an all-or-nothing way. Here, Ghusinga et al. have taken a theoretical kinetic approach to examine how three types of switch architectures affect the properties of the signaling pathway, using a two-tier model. The first tier is the activation of a receptor by a stimulus, denoted as X -> X*. The second tier is the toggling of the switch between inactive (Y) and active (Y*) state, determined by the kinetics of activation and de-activation. In the activation model, the rate of switch activation is correlated with X*; in other words, active receptor turns the switch on. In the derepression model, the rate of switch deactivation is correlated with X; in other words, inactive receptor turns the switch off. The third model, the concerted model, combines activation and derepression. The authors then examine the sensitivity of each switch type to stimulus amount (e.g., receptor occupancy; dose-response curves), how quickly the response changes with increasing stimulus, and sensitivity to fluctuations. The authors describe the consequences and situational values of each type of switch (as an example, the activation model is sensitive and fast but can lead to false positives, making it a good fit for the flight-or-fight adrenaline response). By contrast, slower derepression signaling is more common in resource-limited plants. This analysis will contribute the development of quantitative models of plants. (Summary by Mary Williams @PlantTeaching) Proc. Natl. Acad. Sci. USA 10.1073/pnas.2013401118