Metabolism gene prediction using diversiform molecular features (PNAS)
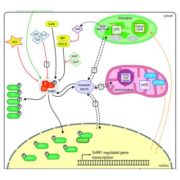
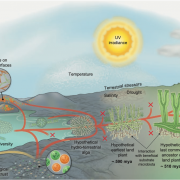
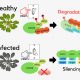
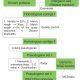
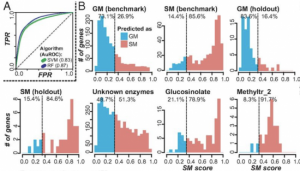 Advances in sequencing technologies enable scientists to obtain molecular features of genes in high-dimensionality. Features of individual gene like expression, methylation, histone modification, evolutionary signals and sequence itself provide high resolution for distinguishing annotated genes. In plant genomes the percentage of annotated genes with experimental evidence is low. In Moore et al., the authors classified Arabidopsis annotated genes as Specialized Metabolism (SM) or General Metabolism (GM). With the integration of 10,243 features, authors conducted machine learning algorithms, Random Forest and Support Vector Machine, to make predictions on identifying genes involved in GM or SM, resulting in “a prediction model … with a true positive rate of 87% and a true negative rate of 71%,” and also identified many genes with unknown function as having potential roles in SM. The study provides a framework on gene function prediction using accessible features from molecular level. (Summary by Zhikai Liang) PNAS 10.1073/pnas.1817074116
Advances in sequencing technologies enable scientists to obtain molecular features of genes in high-dimensionality. Features of individual gene like expression, methylation, histone modification, evolutionary signals and sequence itself provide high resolution for distinguishing annotated genes. In plant genomes the percentage of annotated genes with experimental evidence is low. In Moore et al., the authors classified Arabidopsis annotated genes as Specialized Metabolism (SM) or General Metabolism (GM). With the integration of 10,243 features, authors conducted machine learning algorithms, Random Forest and Support Vector Machine, to make predictions on identifying genes involved in GM or SM, resulting in “a prediction model … with a true positive rate of 87% and a true negative rate of 71%,” and also identified many genes with unknown function as having potential roles in SM. The study provides a framework on gene function prediction using accessible features from molecular level. (Summary by Zhikai Liang) PNAS 10.1073/pnas.1817074116