Loop Assembly: a simple and open system for recursive fabrication of DNA circuits (bioRxiv)
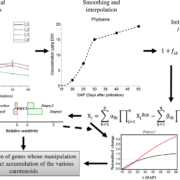
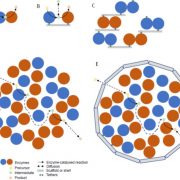
 Ambitious plant synthetic biology projects are driving the evolution of advanced DNA assembly techniques that permit efficient and rapid assembly of multiple transcriptional units (TUs). Pollack et al. have incorporated features of individual techniques into a single “general purpose DNA assembly system”. The system takes advantage of the standardisation of the MoClo/GoldenBraid/PhytoBricks syntax, and uses recursive digestion-ligation cycles (BsaI & SapI) to build plasmids with 16 TUs from the individual parts in three cloning reactions. The plasmids are also compatible with Gibson assembly, providing an alternative assembly pipeline. Cloned plasmids can be propagated in Agrobacterium tumefaciens and are suitable for plant transformation. In conjunction with this system, the authors outline OpenMTA, a framework for the open sharing of DNA parts without restriction on usage or redistribution which includes the Loop Assembly plasmids. In addition, the authors describe LoopDesigner, an open source software for assembly design using Loop Assembly. (Summary by Mike Page) bioRxiv 10.1101/247593
Ambitious plant synthetic biology projects are driving the evolution of advanced DNA assembly techniques that permit efficient and rapid assembly of multiple transcriptional units (TUs). Pollack et al. have incorporated features of individual techniques into a single “general purpose DNA assembly system”. The system takes advantage of the standardisation of the MoClo/GoldenBraid/PhytoBricks syntax, and uses recursive digestion-ligation cycles (BsaI & SapI) to build plasmids with 16 TUs from the individual parts in three cloning reactions. The plasmids are also compatible with Gibson assembly, providing an alternative assembly pipeline. Cloned plasmids can be propagated in Agrobacterium tumefaciens and are suitable for plant transformation. In conjunction with this system, the authors outline OpenMTA, a framework for the open sharing of DNA parts without restriction on usage or redistribution which includes the Loop Assembly plasmids. In addition, the authors describe LoopDesigner, an open source software for assembly design using Loop Assembly. (Summary by Mike Page) bioRxiv 10.1101/247593