JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom
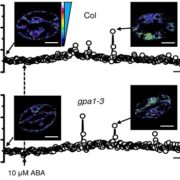
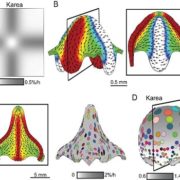
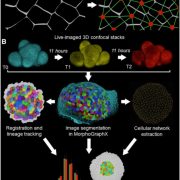
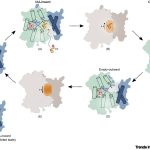
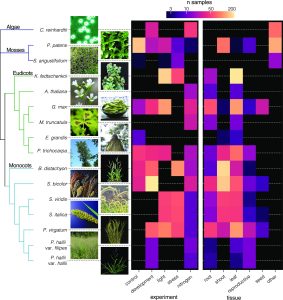 Functional genomic studies across plants are incomplete without deciphering the putative gene functions in model as well as non-model plants. With an aim to facilitate gene function identification and cross-species expression analysis, Sreedasyam et al. describe the Joint Genome Institute (JGI) Plant Gene Atlas, an updateable large-scale data-resource from 18 diverse plant species spanning single celled algae, bryophytes and flowering plants. The JGI Gene Atlas is a data store and analysis platform that contains 15.4 trillion sequenced RNA bases (Tb) from 2090 RNA-Seq data sets collected from 18 plant species. The authors have standardized experimental conditions, tissue types and analytical protocols to facilitate gene prediction across species. The expression profiles of annotated genes were catalogued and co-expression networks were constructed of tissue/condition specific expression patterns, including responses to nitrogen regimes, abiotic stressors, and developmental stages. Along with GO and KEGG pathway enrichment analysis, gene function descriptions are assigned as genes with good (GGF) and genes with poor (GPF) function to expand understanding of gene function in these plant genomes. Apart from discovering enriched gene groups and orthologous groups of genes, the authors provide a framework to discover novel biological role of genes. These datasets are important resources to understand genome annotations in plants, especially future studies related to conserved biological processes. The JGI Plant Gene Atlas Data are currently available at https://plantgeneatlas.jgi.doe.gov and https://phytozome.jgi.doe.gov/ portal. (Summary by Indrani Kakati Baruah, @Indranik333) Nucleic Acids Research (https://doi.org/10.1093/nar/gkad616
Functional genomic studies across plants are incomplete without deciphering the putative gene functions in model as well as non-model plants. With an aim to facilitate gene function identification and cross-species expression analysis, Sreedasyam et al. describe the Joint Genome Institute (JGI) Plant Gene Atlas, an updateable large-scale data-resource from 18 diverse plant species spanning single celled algae, bryophytes and flowering plants. The JGI Gene Atlas is a data store and analysis platform that contains 15.4 trillion sequenced RNA bases (Tb) from 2090 RNA-Seq data sets collected from 18 plant species. The authors have standardized experimental conditions, tissue types and analytical protocols to facilitate gene prediction across species. The expression profiles of annotated genes were catalogued and co-expression networks were constructed of tissue/condition specific expression patterns, including responses to nitrogen regimes, abiotic stressors, and developmental stages. Along with GO and KEGG pathway enrichment analysis, gene function descriptions are assigned as genes with good (GGF) and genes with poor (GPF) function to expand understanding of gene function in these plant genomes. Apart from discovering enriched gene groups and orthologous groups of genes, the authors provide a framework to discover novel biological role of genes. These datasets are important resources to understand genome annotations in plants, especially future studies related to conserved biological processes. The JGI Plant Gene Atlas Data are currently available at https://plantgeneatlas.jgi.doe.gov and https://phytozome.jgi.doe.gov/ portal. (Summary by Indrani Kakati Baruah, @Indranik333) Nucleic Acids Research (https://doi.org/10.1093/nar/gkad616