Plant Science Research Weekly: Sept. 1, 2023
Perspective: How flower development genes were identified using forward genetic screens in Arabidopsis thaliana
 If you weren’t around in the ’80s, you missed some great times; the music, the fashion, the flower genetics! This historical perspective article by David Smythe nicely sums up those heady days with an account of how Arabidopsis thaliana blasted into the ranks of a top model organism, in part through the uncovering of genes controlling flower development. The article describes the steps that occurred in order for these genes to be found and the models of floral development revealed, from chemical mutagenesis of dry seeds, phenotypic screening, backcrossing, and double-mutant analysis. Beyond floral organ identity, these approaches also revealed now-familiar genes involved in auxin transport and meristem maintenance and identity. (Summary by Mary Williams @PlantTeaching) Genetics 10.1093/genetics/iyad102
If you weren’t around in the ’80s, you missed some great times; the music, the fashion, the flower genetics! This historical perspective article by David Smythe nicely sums up those heady days with an account of how Arabidopsis thaliana blasted into the ranks of a top model organism, in part through the uncovering of genes controlling flower development. The article describes the steps that occurred in order for these genes to be found and the models of floral development revealed, from chemical mutagenesis of dry seeds, phenotypic screening, backcrossing, and double-mutant analysis. Beyond floral organ identity, these approaches also revealed now-familiar genes involved in auxin transport and meristem maintenance and identity. (Summary by Mary Williams @PlantTeaching) Genetics 10.1093/genetics/iyad102
Review: Substrate recognition and transport mechanism of the PIN-FORMED auxin exporters
 The PIN-FORMED family of auxin transporters were first identified through genetic screens of Arabidopsis thaliana in the 1980s. Recessive pin mutants form a flowerless, pin-like inflorescence. Subsequent work showed that the encoded genes are membrane-localized auxin transporters and contribute to the export of auxin from the cell. In this review, Ung et al. build on their pivotal 2022 studies and provide a deep look into the structure of PIN transporters. PINs assemble as homodimers, and each monomer includes a scaffold domain and a transporter domain. Particularly interesting is the discussion of how PINs move auxin, through a “cross-over elevator” motif; basically, the substrate is shifted along the transport channel as if in an elevator shaft as the transporter toggles between the Inward and Outward state. The authors also discuss the relatively low affinity of the PINs for auxin, which as they point out means that auxin efflux increases with cytosolic auxin concentration, and also prevents PINs from competing with the very important cytosolic auxin receptors. It’s a fascinating article to add to your reading list. (Summary by Mary Williams @PlantTeaching) TIBS 10.1016/j.tibs.2023.07.006
The PIN-FORMED family of auxin transporters were first identified through genetic screens of Arabidopsis thaliana in the 1980s. Recessive pin mutants form a flowerless, pin-like inflorescence. Subsequent work showed that the encoded genes are membrane-localized auxin transporters and contribute to the export of auxin from the cell. In this review, Ung et al. build on their pivotal 2022 studies and provide a deep look into the structure of PIN transporters. PINs assemble as homodimers, and each monomer includes a scaffold domain and a transporter domain. Particularly interesting is the discussion of how PINs move auxin, through a “cross-over elevator” motif; basically, the substrate is shifted along the transport channel as if in an elevator shaft as the transporter toggles between the Inward and Outward state. The authors also discuss the relatively low affinity of the PINs for auxin, which as they point out means that auxin efflux increases with cytosolic auxin concentration, and also prevents PINs from competing with the very important cytosolic auxin receptors. It’s a fascinating article to add to your reading list. (Summary by Mary Williams @PlantTeaching) TIBS 10.1016/j.tibs.2023.07.006
A chloroplast protein atlas reveals punctate structures and spatial organization of biosynthetic pathways
 Chloroplasts are the location of key processes including photosynthesis, starch synthesis and lipid synthesis. However, many chloroplast proteins have unknown functions, a problem that can in part by addressed through high-resolution localization data. Here, 1034 putative chloroplast- localized proteins were tagged with a C-terminal fluorescent Venus tag, constitutively expressed in the green alga Chlamydomonas reinhardtii, followed by protein localization using confocal microscopy. 28 proteins with known localizations were used to trial the method. Of these, 27 localizations matched previous results, suggesting this method has a high accuracy. Of the 1034 proteins, 581 localized to distinct chloroplast subcompartments, with the pattern of localization often providing a clue to protein function. Interestingly, some enzymes, including those in the chlorophyll biosynthetic pathways, localized to puncta that do not correspond to any chloroplast structure. These puncta might concentrate an enzyme to where its substrate is plentiful or regulate a specific reaction. If you are wondering about the localization of your favorite protein, the authors made their localization data available online and also integrated it into a protein prediction program called PB-Chlamy, both valuable resources to the wider community. (Summary by Rose McNelly @Rose_McN) Cell 10.1016/j.cell.2023.06.008
Chloroplasts are the location of key processes including photosynthesis, starch synthesis and lipid synthesis. However, many chloroplast proteins have unknown functions, a problem that can in part by addressed through high-resolution localization data. Here, 1034 putative chloroplast- localized proteins were tagged with a C-terminal fluorescent Venus tag, constitutively expressed in the green alga Chlamydomonas reinhardtii, followed by protein localization using confocal microscopy. 28 proteins with known localizations were used to trial the method. Of these, 27 localizations matched previous results, suggesting this method has a high accuracy. Of the 1034 proteins, 581 localized to distinct chloroplast subcompartments, with the pattern of localization often providing a clue to protein function. Interestingly, some enzymes, including those in the chlorophyll biosynthetic pathways, localized to puncta that do not correspond to any chloroplast structure. These puncta might concentrate an enzyme to where its substrate is plentiful or regulate a specific reaction. If you are wondering about the localization of your favorite protein, the authors made their localization data available online and also integrated it into a protein prediction program called PB-Chlamy, both valuable resources to the wider community. (Summary by Rose McNelly @Rose_McN) Cell 10.1016/j.cell.2023.06.008
Branched chain amino acids accumulation contributes to drought tolerance in rice
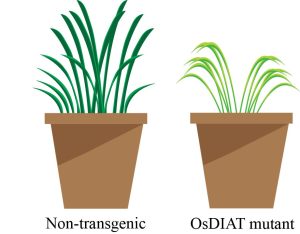 The accumulation of metabolites such as proline as compatible osmolytes is a protective mechanism against drought stress in plants. Similarly, under drought conditions, there is an observable increase in the accumulation of branched-chain amino acids (BCAAs), including the essential amino acids isoleucine, leucine, and valine. Shim et al. investigated rice OsDIAT (DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE), a pivotal enzyme involved in BCAA biosynthesis and the final transamination of BCAAs. RNA-sequencing and gene expression analysis showed that OsDIAT was significantly upregulated under drought conditions. Through CRISPR gene editing, the researchers engineered osdiat mutants, which showed heightened susceptibility to drought and reduced accumulation of BCAAs. Conversely, overexpressing OsDIAT conferred higher plant survival rates and yield under drought conditions. BCAA content in leaves due to the overexpression of OsDIAT was significantly higher under normal condition and further increased upon the exposure to drought when compared with non-transgenic plants. The results suggest that OsDIAT regulates the accumulation of BCAAs in response to drought, which confers drought tolerance in plants. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Physiol. 10.1093/plphys/kiac560
The accumulation of metabolites such as proline as compatible osmolytes is a protective mechanism against drought stress in plants. Similarly, under drought conditions, there is an observable increase in the accumulation of branched-chain amino acids (BCAAs), including the essential amino acids isoleucine, leucine, and valine. Shim et al. investigated rice OsDIAT (DROUGHT-INDUCED BRANCHED-CHAIN AMINO ACID AMINOTRANSFERASE), a pivotal enzyme involved in BCAA biosynthesis and the final transamination of BCAAs. RNA-sequencing and gene expression analysis showed that OsDIAT was significantly upregulated under drought conditions. Through CRISPR gene editing, the researchers engineered osdiat mutants, which showed heightened susceptibility to drought and reduced accumulation of BCAAs. Conversely, overexpressing OsDIAT conferred higher plant survival rates and yield under drought conditions. BCAA content in leaves due to the overexpression of OsDIAT was significantly higher under normal condition and further increased upon the exposure to drought when compared with non-transgenic plants. The results suggest that OsDIAT regulates the accumulation of BCAAs in response to drought, which confers drought tolerance in plants. (Summary by Kumanan N Govaichelvan, @NGKumanan) Plant Physiol. 10.1093/plphys/kiac560
Compartmentation of photosynthesis gene expression in C4 maize depends on time of day
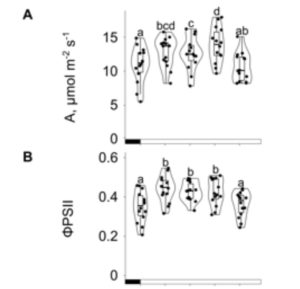 C4 photosynthesis involves the compartmentalization of the photosynthetic reactions across bundle sheath and mesophyll cells, which requires cell-specific gene expression; however, we do not fully understand how this occurs or how photoperiod impacts it. Here maize (Zea mays) was grown in constant light/temperature conditions, and chlorophyll fluorescence and carbon assimilation were measured over 14-hours. The carbon assimilation rate increased from 0 to 10 hours reaching its maximum 10 hours after dawn, whereas maximal photosystem II efficiency occurred 2 hours after dawn. The lack of synchronization between the maximal carbon assimilation rate and photosystem II efficiency might be due to the saturation level of the carbon concentration mechanism, or which decarboxylase is functioning. To further investigate cell specific gene expression, the authors extracted and sequenced RNA from mesophyll and bundle sheath cells over 24-hours. A principal component analysis revealed that both the time of day and cell type affect transcript levels. The largest differences occurred when carbon fixation was at its maximum, 6-10 hrs after dawn, with over 12,000 differentially expressed transcripts. Therefore, this work sheds insight into how the time-of-day influences cell specific gene expression in C4 maize. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad447
C4 photosynthesis involves the compartmentalization of the photosynthetic reactions across bundle sheath and mesophyll cells, which requires cell-specific gene expression; however, we do not fully understand how this occurs or how photoperiod impacts it. Here maize (Zea mays) was grown in constant light/temperature conditions, and chlorophyll fluorescence and carbon assimilation were measured over 14-hours. The carbon assimilation rate increased from 0 to 10 hours reaching its maximum 10 hours after dawn, whereas maximal photosystem II efficiency occurred 2 hours after dawn. The lack of synchronization between the maximal carbon assimilation rate and photosystem II efficiency might be due to the saturation level of the carbon concentration mechanism, or which decarboxylase is functioning. To further investigate cell specific gene expression, the authors extracted and sequenced RNA from mesophyll and bundle sheath cells over 24-hours. A principal component analysis revealed that both the time of day and cell type affect transcript levels. The largest differences occurred when carbon fixation was at its maximum, 6-10 hrs after dawn, with over 12,000 differentially expressed transcripts. Therefore, this work sheds insight into how the time-of-day influences cell specific gene expression in C4 maize. (Summary by Rose McNelly @Rose_McN) Plant Physiol. 10.1093/plphys/kiad447
Plant secondary metabolite-dependent plant-soil feedbacks can improve crop yield in the field
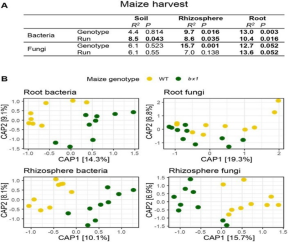 Plant secondary metabolites like benzoxazinoid can alter biotic and abiotic soil properties to shape plant performance through plant-soil feedback, but how this feedback affects agricultural productivity and food quality in the field context is unknown. Here, the authors used large-scale agricultural field experiments to assess the performance of winter wheat varieties growing in field plots whose soils had been conditioned by either wild-type or benzoxazinoid-deficient bx1 maize mutant plants. Results indicated that wheat yield was increased by over 4% without a reduction in grain quality in benzoxazinoid-conditioned soils. The higher plant biomass and kernel yield resulted from the increased subsequent wheat emergence, tillering, and plant performance in the field. The influence of the benzoxazinoid was also observed in the chemical and microbial fingerprints, although the former was more long-lived compared to the latter. This study provides evidence that soil conditioning by plant secondary metabolite-producing plants can increase crop yield via plant-soil feedback under agronomically realistic conditions. Future studies will be important to explore the generalizability and applicability of this concept. (Summary by Abdulmalik, Abdulkabir Omeiza @Omeiza_PlantDoc) eLIFE 10.7554/eLife.84988
Plant secondary metabolites like benzoxazinoid can alter biotic and abiotic soil properties to shape plant performance through plant-soil feedback, but how this feedback affects agricultural productivity and food quality in the field context is unknown. Here, the authors used large-scale agricultural field experiments to assess the performance of winter wheat varieties growing in field plots whose soils had been conditioned by either wild-type or benzoxazinoid-deficient bx1 maize mutant plants. Results indicated that wheat yield was increased by over 4% without a reduction in grain quality in benzoxazinoid-conditioned soils. The higher plant biomass and kernel yield resulted from the increased subsequent wheat emergence, tillering, and plant performance in the field. The influence of the benzoxazinoid was also observed in the chemical and microbial fingerprints, although the former was more long-lived compared to the latter. This study provides evidence that soil conditioning by plant secondary metabolite-producing plants can increase crop yield via plant-soil feedback under agronomically realistic conditions. Future studies will be important to explore the generalizability and applicability of this concept. (Summary by Abdulmalik, Abdulkabir Omeiza @Omeiza_PlantDoc) eLIFE 10.7554/eLife.84988
Functional diversification of a wild potato immune receptor at its center of origin
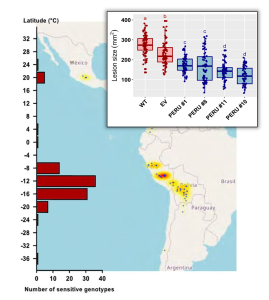 In many biology textbooks, plant pathology is introduced through the historical context of the 1840s great potato famine, caused by colonial ideologies and a virulent pathogen. This causal agent, Phytophthora infestans, is an oomycete that is still present in the environment and causing outbreaks of late blight in solanaceous plants. Here, new insights into the molecular interactions between Phytophthora and potato are uncovered, starting from the discovery in 1994 of Pep-13, an oomycete-produced immunogenic peptide. Screens in wild potato accessions eventually led to the identification of a receptor protein that recognizes Pep-13, named Pep-13 receptor unit (PERU; an excellent name that reflects the geographic center of origin of potato). PERU is a leucine-rich repeat receptor kinase (LRR) protein, similar to other pathogen-recognition proteins. The authors found the greatest number and diversity of PERU alleles in accessions from the Central Andes (Peru), and also found evidence that PERU shows functional diversification in these populations of tuber-bearing Solanum. This elegant research not only adds a nice chapter to the potato / Phytophthora story, but also provides a new tool in the fight against late blight. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adg5261
In many biology textbooks, plant pathology is introduced through the historical context of the 1840s great potato famine, caused by colonial ideologies and a virulent pathogen. This causal agent, Phytophthora infestans, is an oomycete that is still present in the environment and causing outbreaks of late blight in solanaceous plants. Here, new insights into the molecular interactions between Phytophthora and potato are uncovered, starting from the discovery in 1994 of Pep-13, an oomycete-produced immunogenic peptide. Screens in wild potato accessions eventually led to the identification of a receptor protein that recognizes Pep-13, named Pep-13 receptor unit (PERU; an excellent name that reflects the geographic center of origin of potato). PERU is a leucine-rich repeat receptor kinase (LRR) protein, similar to other pathogen-recognition proteins. The authors found the greatest number and diversity of PERU alleles in accessions from the Central Andes (Peru), and also found evidence that PERU shows functional diversification in these populations of tuber-bearing Solanum. This elegant research not only adds a nice chapter to the potato / Phytophthora story, but also provides a new tool in the fight against late blight. (Summary by Mary Williams @PlantTeaching) Science 10.1126/science.adg5261
JGI Plant Gene Atlas: an updateable transcriptome resource to improve functional gene descriptions across the plant kingdom
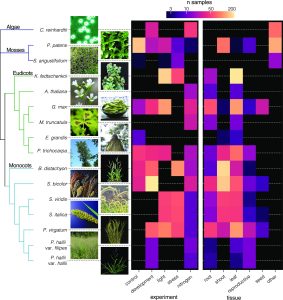 Functional genomic studies across plants are incomplete without deciphering the putative gene functions in model as well as non-model plants. With an aim to facilitate gene function identification and cross-species expression analysis, Sreedasyam et al. describe the Joint Genome Institute (JGI) Plant Gene Atlas, an updateable large-scale data-resource from 18 diverse plant species spanning single celled algae, bryophytes and flowering plants. The JGI Gene Atlas is a data store and analysis platform that contains 15.4 trillion sequenced RNA bases (Tb) from 2090 RNA-Seq data sets collected from 18 plant species. The authors have standardized experimental conditions, tissue types and analytical protocols to facilitate gene prediction across species. The expression profiles of annotated genes were catalogued and co-expression networks were constructed of tissue/condition specific expression patterns, including responses to nitrogen regimes, abiotic stressors, and developmental stages. Along with GO and KEGG pathway enrichment analysis, gene function descriptions are assigned as genes with good (GGF) and genes with poor (GPF) function to expand understanding of gene function in these plant genomes. Apart from discovering enriched gene groups and orthologous groups of genes, the authors provide a framework to discover novel biological role of genes. These datasets are important resources to understand genome annotations in plants, especially future studies related to conserved biological processes. The JGI Plant Gene Atlas Data are currently available at https://plantgeneatlas.jgi.doe.gov and https://phytozome.jgi.doe.gov/ portal. (Summary by Indrani Kakati, @Indranik333) Nucleic Acids Research (https://doi.org/10.1093/nar/gkad616
Functional genomic studies across plants are incomplete without deciphering the putative gene functions in model as well as non-model plants. With an aim to facilitate gene function identification and cross-species expression analysis, Sreedasyam et al. describe the Joint Genome Institute (JGI) Plant Gene Atlas, an updateable large-scale data-resource from 18 diverse plant species spanning single celled algae, bryophytes and flowering plants. The JGI Gene Atlas is a data store and analysis platform that contains 15.4 trillion sequenced RNA bases (Tb) from 2090 RNA-Seq data sets collected from 18 plant species. The authors have standardized experimental conditions, tissue types and analytical protocols to facilitate gene prediction across species. The expression profiles of annotated genes were catalogued and co-expression networks were constructed of tissue/condition specific expression patterns, including responses to nitrogen regimes, abiotic stressors, and developmental stages. Along with GO and KEGG pathway enrichment analysis, gene function descriptions are assigned as genes with good (GGF) and genes with poor (GPF) function to expand understanding of gene function in these plant genomes. Apart from discovering enriched gene groups and orthologous groups of genes, the authors provide a framework to discover novel biological role of genes. These datasets are important resources to understand genome annotations in plants, especially future studies related to conserved biological processes. The JGI Plant Gene Atlas Data are currently available at https://plantgeneatlas.jgi.doe.gov and https://phytozome.jgi.doe.gov/ portal. (Summary by Indrani Kakati, @Indranik333) Nucleic Acids Research (https://doi.org/10.1093/nar/gkad616