Initiation of meiotic recombination is regulated by epigenetic marks and chromatin structure (Genome Res.)
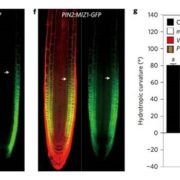
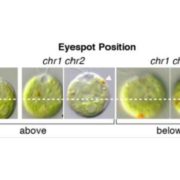
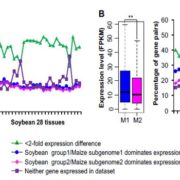
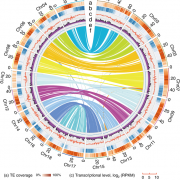
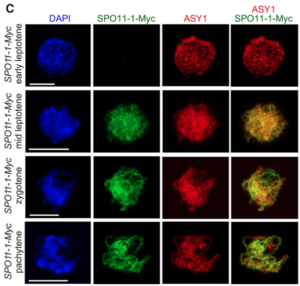 Meiotic recombination is an important source of genetic diversity by allowing the reshuffling of parental alleles during meiosis. It requires the generation of DNA double-strand breaks (DSBs) initiated by the activity of the SPO11 protein complex. In plants, the impact of chromatin structure and DNA methylation on the meiotic DSB landscape genome-wide remains unclear. In this study, the authors sequenced SPO11-oligonucleotides – short DNA sequences that are a by-product of SPO11 activity and indicative of the DSB location – to provide a high-resolution map of meiotic DSB in Arabidopsis thaliana. They show that meiotic DSBs hotspots preferentially occur in gene promoter and terminator nucleosome-free regions, but also in DNA transposons such as Helitrons and MuDR that show low nucleosome occupancy. Transposons are usually heavily transcriptionally silenced by epigenetic mechanisms such as DNA methylation. To investigate the effect of DNA methylation on the meiotic DSB landscape, the authors sequenced SPO11-oligos in the DNA methylation mutant met1. Remarkably, they observed an increase in meiotic DSB hotspots at pericentromeric transposons compared to wild-type, associated with a reduction in nucleosomal occupancy and activation of transcription. Altogether, this study provides new insights in the interplay between chromatin structure, genes and transposons in the regulation of meiotic DSBs, with important consequences on genetic diversity and genome evolution. (Summary by Matthias Benoit) Genome Research 10.1101/gr.225599.117
Meiotic recombination is an important source of genetic diversity by allowing the reshuffling of parental alleles during meiosis. It requires the generation of DNA double-strand breaks (DSBs) initiated by the activity of the SPO11 protein complex. In plants, the impact of chromatin structure and DNA methylation on the meiotic DSB landscape genome-wide remains unclear. In this study, the authors sequenced SPO11-oligonucleotides – short DNA sequences that are a by-product of SPO11 activity and indicative of the DSB location – to provide a high-resolution map of meiotic DSB in Arabidopsis thaliana. They show that meiotic DSBs hotspots preferentially occur in gene promoter and terminator nucleosome-free regions, but also in DNA transposons such as Helitrons and MuDR that show low nucleosome occupancy. Transposons are usually heavily transcriptionally silenced by epigenetic mechanisms such as DNA methylation. To investigate the effect of DNA methylation on the meiotic DSB landscape, the authors sequenced SPO11-oligos in the DNA methylation mutant met1. Remarkably, they observed an increase in meiotic DSB hotspots at pericentromeric transposons compared to wild-type, associated with a reduction in nucleosomal occupancy and activation of transcription. Altogether, this study provides new insights in the interplay between chromatin structure, genes and transposons in the regulation of meiotic DSBs, with important consequences on genetic diversity and genome evolution. (Summary by Matthias Benoit) Genome Research 10.1101/gr.225599.117