High-temporal-resolution Transcriptome Landscape of Early Maize Seed Development
https://doi.org/10.1105/tpc.18.00961
Yi et al. describe a high-temporal-resolution transcriptome landscape of early maize seed development.
By Fei Yi, Wei Gu, and Jinsheng Lai, State Key Laboratory of Agrobiotechnology and National Maize Improvement Center, Department of Plant Genetics and Breeding, China Agricultural University, Beijing, 100193, P. R. China
Background:
Maize seed is an important source of food, feed and biofuel materials. The early maize seed undergoes several developmental stages after double fertilization to become fully differentiated within a short period of time, but the genetic control of this highly dynamic and complex developmental processes remains largely unknown. Understanding the spatial and temporal gene expressional profile along seed development is useful for unraveling the genetic control of seed development and thus for the genetic improvement of this important crop.
Question: We wanted to know the gene activity dynamic during double fertilization, coenocyte formation, cellularization, and differentiation, four main stages of early maize seed development, especially, which genes are specifically expressed at particular stages of early maize seed development.
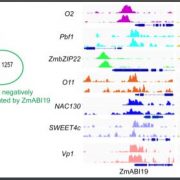
Findings: A total of 22,790 expressed genes including 1,415 transcription factors (TFs) were detected in early stages of maize seed development. In particular, 1,093 genes including 110 TFs were specifically expressed in the seed, most of which were newly identified in this study and displayed high temporal specificity by expressing only in particular period of early seed development. There were 160, 22, 112 and 569 seed-specific genes predominantly expressed in the first 16 hours after pollination, coenocyte formation, cellularization and differentiation stage, respectively. In addition, network analysis predicted 31,256 interactions among 1,317 TFs and 14,540 genes. The high-temporal-resolution transcriptome atlas reported here provides an important resource for future functional study to dissect the genetic control of seed development.
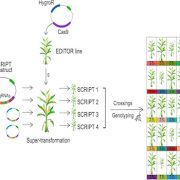
Next steps: We plan to select some key genes for CRISPR/Cas9-based gene editing to further explore their function in the genetic control of seed development.
Fei Yi, Wei Gu, Jian Chen, Ning Song, Xiang Gao, Xiangbo Zhang, Yingsi Zhou, Xuxu M1, Weibin Song, Haiming Zhao, Eddi Esteban, Asher Pasha, Nicholas J. Provart, and Jinsheng Lai (2019). High-temporal-resolution Transcriptome Landscape of Early Maize Seed Development https://doi.org/10.1105/tpc.18.00961
Key words: Maize, seed development, transcriptome, seed-specific genes