Haplotype-resolved genome analyses of a heterozygous diploid potato (Nature Genetics)
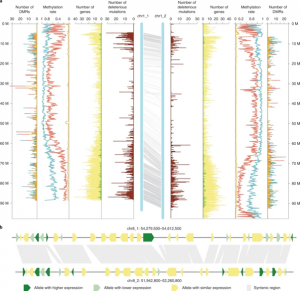 Potato (Solanum tuberosum) is the third most important food crop worldwide. It is also a clonally propagated tetraploid, making progress in breeding genetically improved cultivars extremely difficult. Therefore, there are ongoing efforts to redomesticate potato from a tetraploid, tuber-propagated crop into one that is diploid, inbred-line based, and propagated by seed. However, to do so an improved understanding of the genome landscape of potato is essential. Here, Zhou et al. used various cutting-edge sequencing technologies and a selfing population to generate a haplotype-resolved assembly of a diploid potato. Haplotypes are the distinct sequences of the haploid chromosomes. Most genome assemblies consist of an artificial consensus of both haplotypes, whereas a haplotype-resolved genome considers the different alleles that exist for the same genes, allowing for allele-specific analysis. The authors used two approaches to de novo assemble the potato genome, and eventually combined the results to generate a comprehensive genome of 24 pseudochromosomes. Deleterious mutations and tightly linked genes were identified that will be important targets for future gene editing or molecular selection approaches in this important species. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0699-x
Potato (Solanum tuberosum) is the third most important food crop worldwide. It is also a clonally propagated tetraploid, making progress in breeding genetically improved cultivars extremely difficult. Therefore, there are ongoing efforts to redomesticate potato from a tetraploid, tuber-propagated crop into one that is diploid, inbred-line based, and propagated by seed. However, to do so an improved understanding of the genome landscape of potato is essential. Here, Zhou et al. used various cutting-edge sequencing technologies and a selfing population to generate a haplotype-resolved assembly of a diploid potato. Haplotypes are the distinct sequences of the haploid chromosomes. Most genome assemblies consist of an artificial consensus of both haplotypes, whereas a haplotype-resolved genome considers the different alleles that exist for the same genes, allowing for allele-specific analysis. The authors used two approaches to de novo assemble the potato genome, and eventually combined the results to generate a comprehensive genome of 24 pseudochromosomes. Deleterious mutations and tightly linked genes were identified that will be important targets for future gene editing or molecular selection approaches in this important species. (Summary by Caroline Dowling @CarolineD0wling) Nature Genetics 10.1038/s41588-020-0699-x