Genome-edited rice deficient in two 4CL genes display diverse lignin alterations (Plant Physiol)
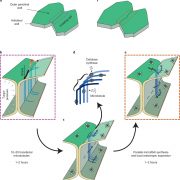
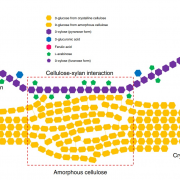
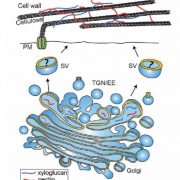
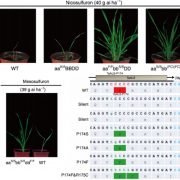
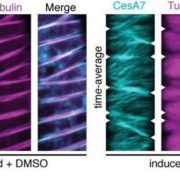
 Lignin is one of the most important end-products of the cinnamate/monolignol pathway and it is abundant in the secondary cell wall of vascular plants. In grasses, lignins are derived from monolignols, p-hydroxycinnamates, and a flavonoid tricin. In the proposed cinnamate/monolignol pathway, 4-COUMARATE:COENZYME A LIGASE (4CL) modulates the pathway flux that converts hydroxycinnamic acids (such as p-coumaric acid) into the products. In the rice genome there are five 4CL isoforms, of which Os4CL3 and Os4CL4 have been suggested to be more relevant than other 4CLs in lignin biosynthesis. In this work, Afifi et al. describe how Os4CL3 and Os4CL4 isoforms have overlapping but divergent roles during the production of lignin monomers in rice. They generated different CRIPSR/Cas9 rice mutant lines for Os4CL3 and Os4CL4 lines and then they analyzed the different cell wall chemical structures. The results suggest that while Os4CL3 might be involved mainly in the production of guaiacyl units and to a lesser extent in the syringyl units, Os4CL4 alone plays a role in tricin biosynthesis. While both mutant lines presented deficiencies in lignin polymer units, each line showed specific characteristics regarding the production of lignin. Concluding, this work proposes new targets for breeding to optimize grass biomass for the sustainable production of biochemicals. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac450
Lignin is one of the most important end-products of the cinnamate/monolignol pathway and it is abundant in the secondary cell wall of vascular plants. In grasses, lignins are derived from monolignols, p-hydroxycinnamates, and a flavonoid tricin. In the proposed cinnamate/monolignol pathway, 4-COUMARATE:COENZYME A LIGASE (4CL) modulates the pathway flux that converts hydroxycinnamic acids (such as p-coumaric acid) into the products. In the rice genome there are five 4CL isoforms, of which Os4CL3 and Os4CL4 have been suggested to be more relevant than other 4CLs in lignin biosynthesis. In this work, Afifi et al. describe how Os4CL3 and Os4CL4 isoforms have overlapping but divergent roles during the production of lignin monomers in rice. They generated different CRIPSR/Cas9 rice mutant lines for Os4CL3 and Os4CL4 lines and then they analyzed the different cell wall chemical structures. The results suggest that while Os4CL3 might be involved mainly in the production of guaiacyl units and to a lesser extent in the syringyl units, Os4CL4 alone plays a role in tricin biosynthesis. While both mutant lines presented deficiencies in lignin polymer units, each line showed specific characteristics regarding the production of lignin. Concluding, this work proposes new targets for breeding to optimize grass biomass for the sustainable production of biochemicals. (Summary by Eva Maria Gomez Alvarez, @eva_ga96) Plant Physiol. 10.1093/plphys/kiac450