Gene Dosage Balance Immediately Following Whole-Genome Duplication in Arabidopsis thaliana
Flowering plants have witnessed multiple cycles of whole-genome duplication (WGD) over the past 200 million years of evolution. Typically, WGD increases genome size and gene content, followed by gene loss, or fractionation, depending on functional categories. Certain classes of genes are retained as duplicates for longer periods of evolutionary time. Duplicate genes provide a potential source of genome diversity and evolution of novel traits (Panchy et al., 2016). Comparative analysis of available genome sequence data provides valuable resources to understand gene duplication and the mechanisms behind duplicate gene retention. Genes involved in macromolecular complexes such as signal transduction and transcription factors are preferentially retained as duplicates, whereas these same gene classes are underrepresented in small-scale duplications (SSD, e.g., tandem duplication, transposition). This pattern of reciprocal retention of gene duplicates originating from WGD signifies the involvement of genomic balance in the evolutionary fate of duplicated genes (Figure).
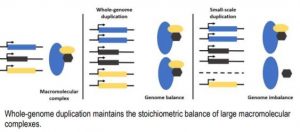 The gene balance hypothesis (GBH) predicts that altering the stoichiometric balance of members of macromolecular complexes leads to dosage-dependent phenotypes (Veitia et al. 2008). WGD overcomes this disadvantage by duplicating every gene in the complex and thus preserving the balance. Although considerable interest exists in understanding the gene dosage responses, very little is known about the gene dosage response to immediate autopolyploidy. In new work, Song et al. (2020) sought to determine the instantaneous effect of gene duplication in the absence of hybridization. Previous work by the authors showed that reciprocally retained genes in Glycine neoallopolyploids displayed a coordinated dosage response compared to non-reciprocally retained genes (Coate et al. 2016). However, the observed gene expression changes were confounded by the combination of WGD, hybridization, and 0.5 million years of evolution post-WGD. To avoid these confounding effects, Song et al. (2020) used diploid/synthetic autotetraploid pairs of Arabidopsis thaliana ecotype C24 and Ws (colchicine-induced tetraploid of the same accessions) and a tetraploid/synthetic diploid pair of the ecotype Wa (diploid generated by Tailswap haploid induction).
The gene balance hypothesis (GBH) predicts that altering the stoichiometric balance of members of macromolecular complexes leads to dosage-dependent phenotypes (Veitia et al. 2008). WGD overcomes this disadvantage by duplicating every gene in the complex and thus preserving the balance. Although considerable interest exists in understanding the gene dosage responses, very little is known about the gene dosage response to immediate autopolyploidy. In new work, Song et al. (2020) sought to determine the instantaneous effect of gene duplication in the absence of hybridization. Previous work by the authors showed that reciprocally retained genes in Glycine neoallopolyploids displayed a coordinated dosage response compared to non-reciprocally retained genes (Coate et al. 2016). However, the observed gene expression changes were confounded by the combination of WGD, hybridization, and 0.5 million years of evolution post-WGD. To avoid these confounding effects, Song et al. (2020) used diploid/synthetic autotetraploid pairs of Arabidopsis thaliana ecotype C24 and Ws (colchicine-induced tetraploid of the same accessions) and a tetraploid/synthetic diploid pair of the ecotype Wa (diploid generated by Tailswap haploid induction).
RNA-seq analysis of the nascent autopolyploids facilitated the quantification of the overall mRNA transcriptome size, variance in gene expression, and gene dosage responses. The authors found that doubling the genome increased the total amount of transcripts per cell but not exactly two-fold. Synthetic tetraploids of C24 and Ws accessions exhibited a small deviation in mRNA transcriptome size per genome relative to their respective diploid progenitors. While C24 showed a reduction in transcriptome size per genome, Ws and the natural tetraploid Wa showed more transcripts per genome. Estimation of absolute dosage responses at individual loci through quantification of transcriptome size showed that changes in transcripts per gene copy were highly variable, and the expression of many genes did not strictly correlate with gene copy number. Reciprocally retained genes that are putatively dosage-sensitive displayed a more coordinated dosage response immediately after WGD.
By leveraging this unique study system, Song et al. were able to examine the transcriptional response immediately following genome doubling. The fact that there was not a more pronounced coordinated dosage response among reciprocally retained genes in nascent polyploids compared to natural allotetraploid (Coate et al. 2016) suggests that the coordination occurs immediately and is maintained. Such coordinated regulation of reciprocally retained genes appears to be robust to hybridization and the resulting regulatory changes. Further studies on older polyploids are necessary to resolve the timeline for relaxation of selection on gene dosage.
Sunil K. Kenchanmane Raju
Department of Plant Biology,
Michigan State University
ORCID ID: 0000-0001-8960-094X
REFERENCES
Panchy, N., Lehti-Shiu, M., & Shiu, S. H. (2016). Evolution of gene duplication in plants. Plant Phys, 171: 2294-2316.
Veitia, R. A., Bottani, S., & Birchler, J. A. (2008). Cellular reactions to gene dosage imbalance: genomic, transcriptomic and proteomic effects. Trends Genet, 24: 390-397.
Coate, J. E., Song, M. J., Bombarely, A., & Doyle, J. J. (2016). Expression‐level support for gene dosage sensitivity in three Glycine subgenus Glycine polyploids and their diploid progenitors. New Phytol, 212: 1083-1093.
Song, M. J., Potter, B. I., Doyle, J. J. & Coate, J. E. (2020). Gene Balance Predicts Transcriptional Responses Immediately Following Ploidy Change in Arabidopsis thaliana. Plant Cell. DOI: https://doi.org/10.1105/tpc.19.00832.



