Feasting While Fasting: How Autophagy Helps Maize Survive Carbon Starvation
Most macro-molecular components of plant cells (e.g., proteins, lipids, and even entire organelles) are subject to an ongoing process of recycling to both rejuvenate aging structures and optimize the allocation of cellular nutrients. A major recycling route is autophagy, which occurs under normal conditions but becomes especially prevalent when nutrients become scarce, such as during fixed-carbon (C) or nitrogen (N) starvation (Marshall and Vierstra, 2018). Autophagy is crucial during such limitations because it allows cells to scavenge their own molecular components to obtain the nutrients needed for sustained metabolism and cellular survival.
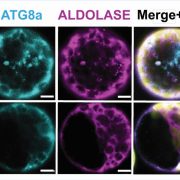
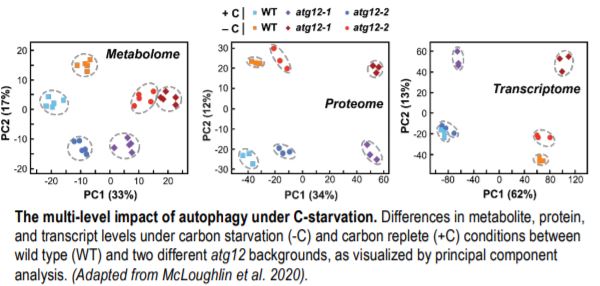 The best understood autophagy process is macroautophagy (hereafter called autophagy), wherein select cytosolic components are encapsulated in autophagosomes and brought to the vacuole for degradation. In a recent study, McLoughlin et al. (2020) performed a detailed multi-level molecular characterization of autophagy-deficient atg12 maize (Zea mays) leaves under normal and C starvation conditions induced by leaf shading. To enhance correlative power, the authors examined two atg12 mutant alleles that were previously shown to attenuate autophagosome formation to different degrees (i.e., complete vs. partial disruption) (Li et al., 2015). The results complement a partner study that examined these same maize leaves under N starvation (McLoughlin et al., 2018). When analyzed together, these datasets helped distinguish autophagy-related responses that are stress specific versus those that persist generally. For example, the levels of N-rich metabolites, including most amino acids and the ureides allantoin and allantoic acid, specifically accumulated in the atg12 backgrounds under C but not N starvation.
The best understood autophagy process is macroautophagy (hereafter called autophagy), wherein select cytosolic components are encapsulated in autophagosomes and brought to the vacuole for degradation. In a recent study, McLoughlin et al. (2020) performed a detailed multi-level molecular characterization of autophagy-deficient atg12 maize (Zea mays) leaves under normal and C starvation conditions induced by leaf shading. To enhance correlative power, the authors examined two atg12 mutant alleles that were previously shown to attenuate autophagosome formation to different degrees (i.e., complete vs. partial disruption) (Li et al., 2015). The results complement a partner study that examined these same maize leaves under N starvation (McLoughlin et al., 2018). When analyzed together, these datasets helped distinguish autophagy-related responses that are stress specific versus those that persist generally. For example, the levels of N-rich metabolites, including most amino acids and the ureides allantoin and allantoic acid, specifically accumulated in the atg12 backgrounds under C but not N starvation.
To generate detailed insights into autophagy-influenced processes, the authors considered relationships among extensive metabolomic, transcriptomic, and proteomic datasets across treatments and genotypes (see Figure). By comparing the transcriptomic and proteomic datasets, the authors could even identify proteins whose abundances were controlled by autophagy alone versus changes in mRNA levels. They found a strong upregulation of catabolic pathways for starch, amino acids, and nucleotides in the absence of autophagy under fixed-C starvation. This likely occurred because the loss of autophagy removed macromolecule recycling as a source of C nutrition, thus accelerating the depletion of carbon stores caused by a lack of photosynthesis. Predictably, the leaves responded by upregulating other catabolic pathways needed to sustain the pool of respiratory carbon substrates. The proteomic datasets also identified a number of proteins and protein complexes whose abundances increased in the atg12 backgrounds and are thus likely autophagic targets.
Large changes to peroxisome-related components and metabolites were also observed in atg12 samples across treatments. While atg12 leaves accumulated more peroxisomal proteins overall due to the lack of autophagic recycling of this organelle, the authors also observed a shift in the protein profile consistent with a transformation of peroxisomes to glyoxysomes with their signature enzymes malate synthase and isocitrate lyase. Glyoxysomes, which are prevalent in germinating oil-rich seeds, primarily function in b-oxidation that generates acetyl-CoA during fatty acid breakdown. The glyoxylate cycle coverts acetyl-CoA to succinate, which can then enter the mitochondrial TCA cycle, leading to the intriguing proposal that in C-starved atg12 leaves, the glyoxylate cycle supports the formation of respiration substrates using carbon skeletons from various sources, including lipid droplets. However, further research is needed to determine whether the shift in peroxisomal metabolism and the observed accumulation of nitrogen-rich metabolites might also reflect a nutrient recovery strategy as part of an early induction of senescence in darkened atg12 leaves.
Brendan M O’Leary
ARC Centre of Excellence in Plant Energy Biology
University of Western Australia
ORCID: 0000-0002-8770-155X
REFERENCES
Li, F., Chung, T., Pennington, J.G., Federico, M.L., Kaeppler, H.F., Kaeppler, S.M., Otegui, M.S., and Vierstra, R.D. (2015). Autophagic recycling plays a central role in maize nitrogen remobilization. Plant Cell 27, 1389-1408.
Marshall, R.S., and Vierstra, R.D. (2018). Autophagy: the master of bulk and selective recycling. Annu. Rev. Plant Biol. 69, 173-208.
McLoughlin, F., Augustine, R.C., Marshall, R.S., Li, F., Kirkpatrick, L.D., Otegui, M.S., and Vierstra, R.D. (2018). Maize multi-omics reveal roles for autophagic recycling in proteome remodelling and lipid turnover. Nat Plants 4, 1056-1070.