Exploring variation in a key maize regulator of floral form
María Jazmín Abraham-Juárez, Amanda Schrager-Lavelle et. al. explore how variation in a key floral regulator affects floral development, downstream gene expression and protein complex assembly in maize. Plant Cell https://doi.org/10.1105/tpc.20.00300
by Jazmin Abraham-Juarez, Biology Department, University of Massachusetts Amherst, USA, and CONACYT-IPICYT, San Luis Potosi, Mexico
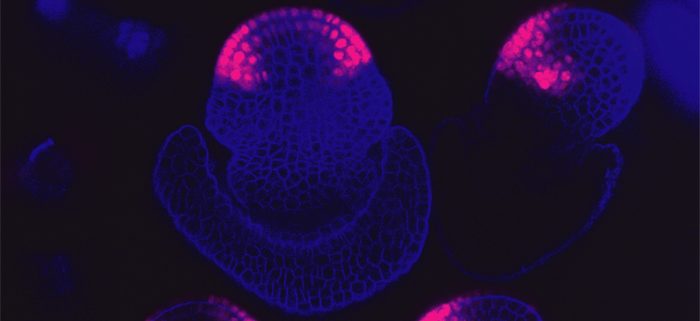
Background. The regulatory network that controls floral organ formation is complex and still not completely understood. This network has also been changing over the course of evolution, leading to immense floral diversity. MADS-box transcription factors are important regulators of floral organ development, and changing interactions between MADS-box proteins are predicted to have influenced floral evolution. However, the functional consequences of evolutionary change to MADS-box protein-protein interactions were unknown. Grasses are an excellent family to explore these consequences due to high genetic diversity, evolutionary variation in MADS-box protein-protein interactions, and the numerous genetic model systems in the family. We used the genetic and genomic resources available in maize to determine the functional consequences of evolutionary variation in dimerization of a key regulator of floral development, STERILE TASSEL SILKY EAR1 (STS1).
Question. STS1 dimerization varies across the grass family. In this work, we set out to discover the effect of some of this evolutionary variation on floral development, downstream gene expression and protein complex assembly in maize.
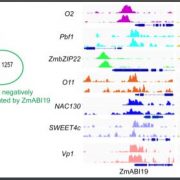
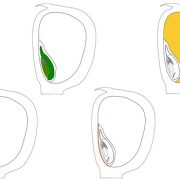
Findings. We found that evolutionary variants of STS1 differentially affect downstream gene expression and protein complex assembly in maize. Surprisingly, we found that STS1 dimerization changed protein degradation dynamics, which had not been described. This discovery adds a new layer of complexity to the regulation of flower development, and how that regulation changes over deep time. We also found that a coding change to STS1, a master regulator of development, had subtle effects on floral development. Our results indicate that small changes to MADS-box genes may have influenced the gradual evolution of floral form. This dissection of the effects of a single base-pair change, using an allelic series provided by evolution, is critical in this era of genome editing.
Next steps. Next projects are focused on explore mechanisms by which protein modifications and interactions affect floral organ formation and evolution. In the future, our findings might help to identify additional factors influencing floral development and evolution.
María Jazmín Abraham-Juárez, Amanda Schrager-Lavelle, Jarrett Man, Clinton Whipple, Pubudu Handakumbura, Courtney Babbitt, Madelaine Bartlett (2020) Evolutionary Variation in MADS-box Dimerization Affects Floral Development and Protein Abundance in Maize. Plant Cell. https://doi.org/10.1105/tpc.20.00300