Expanded function of the P-type pentatricopeptide repeat protein ATP4 in RNA editing
Tianhu Sun
ORCID ID: 0000-0002-2513-1387
Plant Breeding and Genetics Section, School of Integrative Plant Science, Cornell University, Ithaca, New York 14853
Chloroplasts are semi-autonomous organelles that retain their own genomes derived from their cyanobacterial ancestors, but with reduced size and gene number. However, chloroplast genes cannot function autonomously. Instead, nuclear-encoded factors are required for all steps of chloroplast gene expression. These factors promote stabilization, splicing, translation, or editing of RNAs. Therefore, post‐transcriptional control by nuclear-encoded factors is a critical step for chloroplasts to produce functional RNAs and proteins, and finally secure the correct synthesis of the photosynthetic machinery in chloroplasts.
Pentatricopeptide repeat (PPR) proteins are a large family of nuclear-encoded RNA binding proteins involved in RNA metabolism in chloroplasts and mitochondria. The PPR family is subdivided into two groups, P-type and PLS-type, based on their motifs. P-type PPRs mainly function in RNA splicing, stabilization, and translational activation, and PLS-type PPRs are mostly dedicated to RNA editing (Barkan and Small, 2014). RNA editing is an important process in gene regulation and involves nucleotide modification; indeed, many transcripts expressed in chloroplasts are modified by C-to-U RNA editing. However, how plants effectively and precisely edit chloroplast transcripts remains an enigma.
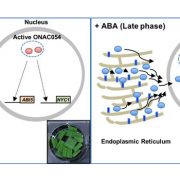
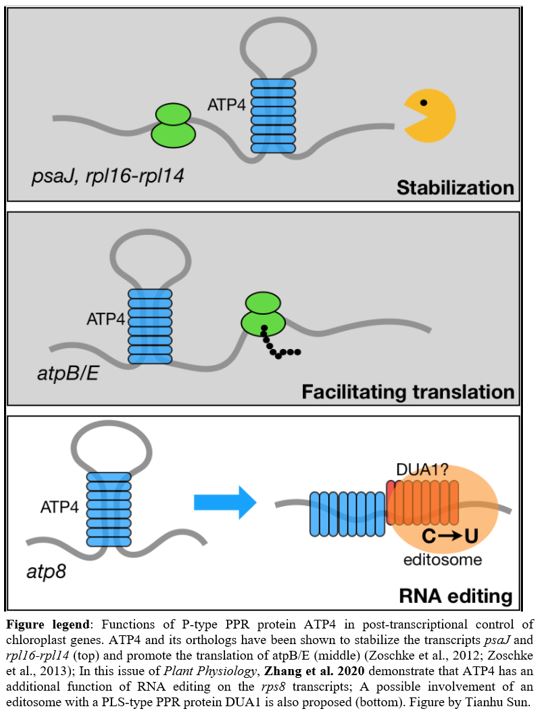 ATP4 (ADENOSINE TRIPHOSPHATASE4) is a P-type PPR protein with a C-terminal MutS-related domain (denoted as an SMR domain; Small MutS Related). ATP4 in maize (Zea mays) and its orthologs (SUPPRESSOR OF VARIEGATION7 (SVR7) in Arabidopsis thaliana and OsPPR676 in rice (Oryza sativa) have been shown to have multiple functions including promoting the translation of atpB/E (Zoschke et al., 2012), stabilizing psaJ and rpl16-rpl14 (Zoschke et al., 2013), and enhancing chloroplast rRNA processing (Liu et al., 2010) (Figure 1). In this issue of Plant Physiology, Zhang et al. (2020) demonstrate that ATP4 in rice and maize has an unanticipated role in editing ribosomal protein S8 transcripts (rps8). Furthermore, they provide genetic evidence that the rps8 expression is responsible for the cold-sensitive phenotype of rice osatp4 mutants.
ATP4 (ADENOSINE TRIPHOSPHATASE4) is a P-type PPR protein with a C-terminal MutS-related domain (denoted as an SMR domain; Small MutS Related). ATP4 in maize (Zea mays) and its orthologs (SUPPRESSOR OF VARIEGATION7 (SVR7) in Arabidopsis thaliana and OsPPR676 in rice (Oryza sativa) have been shown to have multiple functions including promoting the translation of atpB/E (Zoschke et al., 2012), stabilizing psaJ and rpl16-rpl14 (Zoschke et al., 2013), and enhancing chloroplast rRNA processing (Liu et al., 2010) (Figure 1). In this issue of Plant Physiology, Zhang et al. (2020) demonstrate that ATP4 in rice and maize has an unanticipated role in editing ribosomal protein S8 transcripts (rps8). Furthermore, they provide genetic evidence that the rps8 expression is responsible for the cold-sensitive phenotype of rice osatp4 mutants.
Zhang et al. (2020) first characterized a rice osatp4 T-DNA mutant with reduced plant height, tiller number, and chlorophyll content. Similar to the mutation in maize (Zoschke et al., 2012), under natural field conditions the rice osatp4 mutant only affects the accumulation of ATP synthase but no other photosynthetic complexes. By contrast, under cold stress conditions, a global reduction of translation of many chloroplast-encoded genes, including the D1 protein of photosystem II, subunit of cytochrome b6f complex, and the Rubisco large subunit, has been observed in the osatp4 mutant, as well as a significant reduction of chlorophyll content.
The authors then investigated the cause of the global reduction in chloroplast translation. Recently, GUN1 (GENOME UNCOUPLED1), another P-type PPR protein with a SMR domain, was shown to affect chloroplast RNA editing through the interaction with the accessory editing factor MULTIPLE ORGANELLAR RNA EDITING FACTOR 2 (MORF2; Zhao et al., 2019). This work inspired the authors to explore whether ATP4 also influences chloroplast RNA editing. By sequencing chloroplast transcripts with editing sites, Zhang et al. (2020) found that the editing of rps8 is strongly reduced in the osatp4 mutant under both natural field and cold stress conditions. Similarly, the authors found that the maize atp4 mutant showed a defect in rps8 editing. Chloroplast rps8 encodes a protein of the chloroplast small ribosome subunit, so the editing defect of rps8 may explain the global reduction in chloroplast translation.
To address whether the rps8 editing defect is related to the cold-sensitive phenotype in osatp4 mutants, the authors first surveyed the editing efficiency of rps8 in different cultivated rice accessions and found two accessions with no rps8 editing. These two accessions showed a similar albino phenotype to the osatp4 mutant under cold stress conditions. Furthermore, complementation of the osatp4 mutant with the edited form of rps8 can restore a large portion of the chlorophyll accumulation under cold stress conditions. These data indicate that the editing defect of rps8, rather than osatp4 directly, is responsible for the cold-sensitive phenotype. A possible explanation is that RPS8 is essential to maintain high chloroplast translation capacity under cold stress conditions, similar to what was observed for another plastid-encoded ribosomal protein in tobacco (Nicotiana tabacum) (Rogalski et al., 2008).
The next question is, how does ATP4 edit rps8 transcripts? Since P-type PPR proteins lack the motif directly involved in RNA editing and the function of P-type PPR proteins in RNA editing is merely suggested, could it be a secondary effect of the malfunction of other genes in atp4 mutant, such as the defect of rpl16-rpl14 transcription? To address this, Zhang et al. (2020) showed that another maize mutant pgr3, which has a similar rpl16-rpl14 transcript defect to the atp4 mutant, shows normal rps8 editing. This result showed that rpl16-rpl14 transcript accumulation and rps8 editing are two independent events.
In this work, Zhang et al. (2020) clearly demonstrate the expanded function of a P-type PPR protein in RNA editing in addition to the main role of P-type PPR proteins in RNA splicing, stabilization, and translational activation and propose the potential role of RPS8 in rice cold-adaptability. The first report of a P-type PPR protein in RNA editing was PPME in mitochondria (Leu et al., 2016). Recently, the P-type PPR protein GUN1 was also shown to be involved in chloroplast RNA editing (Zhao et al., 2019). Taken together, these studies highlight the broader function of P-type PPR proteins in RNA editing.
It will be interesting to learn more about how ATP4 functions in post-transcriptional control of chloroplast genes. First of all, the function of the SMR domain in ATP4 is still not clear. Furthermore, since ATP4 itself does not contain the domain for C-to-U conversion in RNA, Zhang et al. (2020) proposed that ATP4 may disrupt the stem-loop of rps8, which facilitates the binding of DUA1 (Figure 1), a PLS-type PPR protein that might directly edit rps8 (Cui et al., 2019). Therefore, a hypothetical “editosome” complex, rather than ATP4 itself, may be responsible for RNA editing. The components of the editosome and how they coordinately edit rps8 transcripts need to be further elucidated.
LITERATURE CITED
Barkan A, Small I (2014) Pentatricopeptide repeat proteins in plants. Annu Rev Plant Biol 65: 415-442
Cui X, Wang Y, Wu J, Han X, Gu X, Lu T, Zhang Z (2019) The RNA editing factor DUA 1 is crucial to chloroplast development at low temperature in rice. New Phytol 221: 834-849
Leu K-C, Hsieh M-H, Wang H-J, Hsieh H-L, Jauh G-Y (2016) Distinct role of Arabidopsis mitochondrial P-type pentatricopeptide repeat protein-modulating editing protein, PPME, in nad1 RNA editing. RNA Biol 13: 593-604
Liu X, Yu F, Rodermel S (2010) An Arabidopsis pentatricopeptide repeat protein, SUPPRESSOR OF VARIEGATION7, is required for FtsH-mediated chloroplast biogenesis. Plant Physiol 154: 1588-1601
Rogalski M, Schöttler M A, Thiele W, Schulze W X, Bock R (2008) Rpl33, a nonessential plastid-encoded ribosomal protein in tobacco, is required under cold stress conditions. Plant Cell 20: 2221-2237.
Zhang J, Guo Y, Fang Q, Zhu Y, Zhang Y, Liu X, Lin Y, Barkan A, Zhou F (2020) The PPR-SMR protein ATP4 is required for editing the chloroplast rps8 mRNA in rice and maize. Plant Physiol
Zhao X, Huang J, Chory J (2019) GUN1 interacts with MORF2 to regulate plastid RNA editing during retrograde signaling. Proc Natl Acad Sci USA 116: 10162-10167
Zoschke R, Kroeger T, Belcher S, Schöttler MA, Barkan A, Schmitz‐Linneweber C (2012) The pentatricopeptide repeat‐SMR protein ATP4 promotes translation of the chloroplast atpB/E mRNA. Plant J 72: 547-558
Zoschke R, Watkins KP, Barkan A (2013) A rapid ribosome profiling method elucidates chloroplast ribosome behavior in vivo. Plant Cell 25: 2265-2275