Excising the mystery of single guide RNA processing
Sophia G. Zebell
Howard Hughes Medical Institute, Cold Spring Harbor Laboratory, Cold Spring Harbor, NY, USA
Over the past 10 years, utilization of CRISPR-Cas9 genome editing in plants has rapidly and significantly altered the scale and scope of both basic research and crop development. The transformative nature of genome editing technology has led to a great appetite for optimization. Many researchers have contributed to the development of CRISPR-Cas9 methods, with variable techniques and success rates. There are a number of strategies for eukaryotic deployment of the Cas9 protein and the sgRNA elements, as well as attempts to increase editing efficiency and germline transmission of edited alleles (Mao et al. 2019). One area of focus in this optimization effort is the delivery of multiplexed single guide RNAs (sgRNAs), the single-stranded RNAs that guide the Cas9 enzyme to specific DNA targets.
Clever solutions for sgRNA delivery have included interspersing an array of sgRNAs with flanking self-cleaving ribozymes, tRNAs, or directed-cleavage sequences. Another broadly implemented strategy is to drive nuclear expression of a single mature sgRNA with a U6 promoter, to be transcribed by RNA Polymerase III (Mao et al. 2019). Over time, however, researchers have noticed that these approaches appear to be unnecessary, as delivery of Cas9 and an array of sgRNAs in a single transcriptional unit without any special flanking sequences is just as efficient for Cas9 targeting and generation of DNA double-stranded breaks (Mikami, Toki, and Endo 2017; Wang et al. 2018). This finding pointed strongly to the existence of an unknown mechanism for sgRNA maturation in eukaryotes.
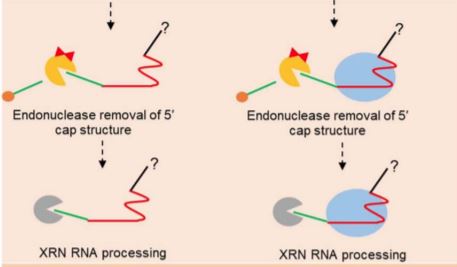
In this issue of Plant Physiology, Cody and Scholthof thoroughly and convincingly demonstrate the presence of native sgRNA processing in plants and present a highly anticipated model for this activity (Cody and Scholthof 2020). Working in vitro, they verified the longstanding assumption that the presence of extra nucleotides 5’ of the sgRNA inhibit Cas9 DNA cleavage. However, in Nicotiana benthamiana sgRNAs transcribed with 5’ overhangs are still functional, and Cas9-sgRNA complexes immunoprecipitated from the plant lack the overhangs, confirming that processing occurs.
To probe the mechanism of 5’ processing, the authors conducted a series of in vitro experiments exposing sgRNA sequences to 5’-3’ exoribonuclease. They found that immature sgRNA containing extra 5’ sequence is readily digested, while mature sgRNA and Cas9-sgRNA complexes immunoprecipitated from plants are inherently resistant to 5’-3’ exoribonuclease activity. These observations led them to suggest two possible models: (1) Cas9 protects the bound sgRNA fragment from exonuclease during processing; and (2) mature sgRNA in vivo resists digestion through unknown 3’ processing or folding mechanisms. These models are not mutually exclusive, and both allow for efficient processing of sgRNAs without the need for encoded signals directing RNA cleavage. They then demonstrated in vitro that the required 5’-3’ exonuclease activity could be accomplished by a member of the XRN family, the predominant 5’-3’ exoribonucleases found in plants and mammals.
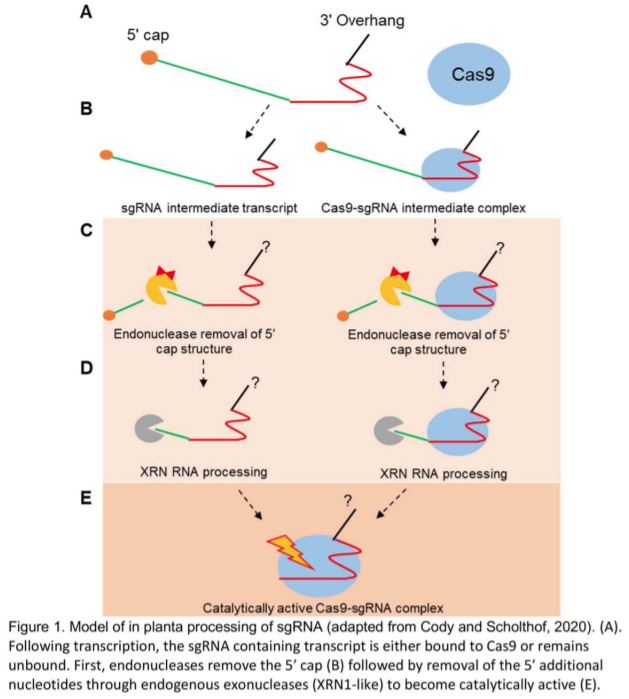 This model (Figure 1) simultaneously offers timely answers to questions about sgRNA processing and highlights the need for further investigation into the mechanism, including genetic studies to identify the precise enzymes responsible for the activities observed. In order to function, XRN exonucleases require a 5’ phosphate group, which implies a need for an unknown endonuclease to initiate sgRNA maturation, either by removing the 5’ cap or by cleaving between an array of guides. Additionally, the authors make no claims about the 3’ maturation of sgRNAs, which will need to be further investigated.
This model (Figure 1) simultaneously offers timely answers to questions about sgRNA processing and highlights the need for further investigation into the mechanism, including genetic studies to identify the precise enzymes responsible for the activities observed. In order to function, XRN exonucleases require a 5’ phosphate group, which implies a need for an unknown endonuclease to initiate sgRNA maturation, either by removing the 5’ cap or by cleaving between an array of guides. Additionally, the authors make no claims about the 3’ maturation of sgRNAs, which will need to be further investigated.
The enthusiasm of plant biologists for CRISPR-Cas9 technology has outpaced our understanding of its in planta mechanism, leaving a gap to be filled by basic science. By demonstrating that eukaryotic processing of sgRNAs could occur through highly conserved enzymatic activities, Cody and Scholthof are encouraging a shift in focus from Cas9 vector engineering to simplified delivery systems and back-to-basics approaches to CRISPR-Cas9. They have also opened the door for optimization of genome editing through enhancing the plant’s capacity to execute editing, which could improve the speed and efficiency of genome editing for model organisms, and possibly help to encourage editing in recalcitrant plant systems.
Literature Cited
Mao, Yanfei, Jose Ramon Botella, Yaoguang Liu, and Jian-Kang Zhu. 2019. “Gene Editing in Plants: Progress and Challenges.” National Science Review 6 (3): 421–37. https://doi.org/10.1093/nsr/nwz005.
Mikami, Masafumi, Seiichi Toki, and Masaki Endo. 2017. “In Planta Processing of the SpCas9–GRNA Complex.” Plant and Cell Physiology 58 (11): 1857–67. https://doi.org/10.1093/pcp/pcx154.
Wang, Mugui, Yanfei Mao, Yuming Lu, Zhidan Wang, Xiaoping Tao, and Jian-Kang Zhu. 2018. “Multiplex Gene Editing in Rice with Simplified CRISPR-Cpf1 and CRISPR-Cas9 Systems.” Journal of Integrative Plant Biology 60 (8): 626–31. https://doi.org/10.1111/jipb.12667.