Design Stars: How GRF-INTERACTING FACTORs Help Determine the Layout of the Root Tip
Roots grow into new regions of the soil in their constant quest for water and dissolved minerals. This crucial task is made possible by the root meristem and stem cell niche, where neatly arranged stem cells quickly divide beneath a protective root cap. Stem cells give rise to transit amplifying cells, which undergo several quick rounds of division before differentiating into the various cells that make up the root. The stem cell region surrounds the quiescent center (QC), comprising only four to seven cells in Arabidopsis thaliana but 500–1200 cells in maize (Zea mays). Unlike stem cells, QC cells normally take more than three days to divide, but they spring into action as needed to replace damaged stem cells (Heyman et al., 2014).
GROWTH-REGULATING FACTORs (GRFs) promote the rapid division of transit amplifying cells while repressing genes that are active in stem cells. GRF activity is enhanced by GRF-INTERACTING FACTORs (GIFs), including ANGUSTIFOLIA3 (AN3)/GIF1, GIF2, and GIF3. Although these small transcriptional activators cannot directly bind to DNA, they increase aerial organ growth in cooperation with GRFs (Horiguchi et al., 2005). However, their roles in root development have been unclear.
Through rigorous analysis of GIF mutants, Ercoli et al. (2018) uncovered vital roles for these small molecules in patterning the Arabidopsis root meristem. The QCs of some GIF mutants such as an3, gif2, gif3, and an3 gif3 appeared normal, whereas an3 gif2 and an3 gif2 gif3 QCs were quite disorganized, suggesting that together, the GIFs help determine QC organization. Primary root growth was normal in some mutants, but others had diverse phenotypes: an3 roots were longer than wild type, whereas an3 gif2 roots were short, and an3 gif2 gif3 roots were even shorter. Despite their impaired QCs, both long- and short-rooted mutants had enlarged meristems due to increased meristem cell numbers. Thus, it appears that GIFs quantitatively control both root meristem size and QC organization in diverse ways. The effects of AN3 were at least partially reversible, as observed in GIF mutants transformed with an inducible AN3 system. AN3, GIF2, and GIF3 had overlapping but distinct expression patterns; only AN3 showed peak expression in the QC and stem cell region.
The authors then asked whether AN3 requires its interacting partners, the GRFs, for its activity. Plants with reduced GRF activity (due to overexpression of the microRNA miR396) had large root meristems but normal QCs, as did plants overexpressing miR396 in the an3 background, while plants overexpressing both miR396 and AN3 had large root meristems with normal QCs. These findings suggest that AN3 controls root meristem length together with the GRFs, but only AN3 helps maintain the QC.
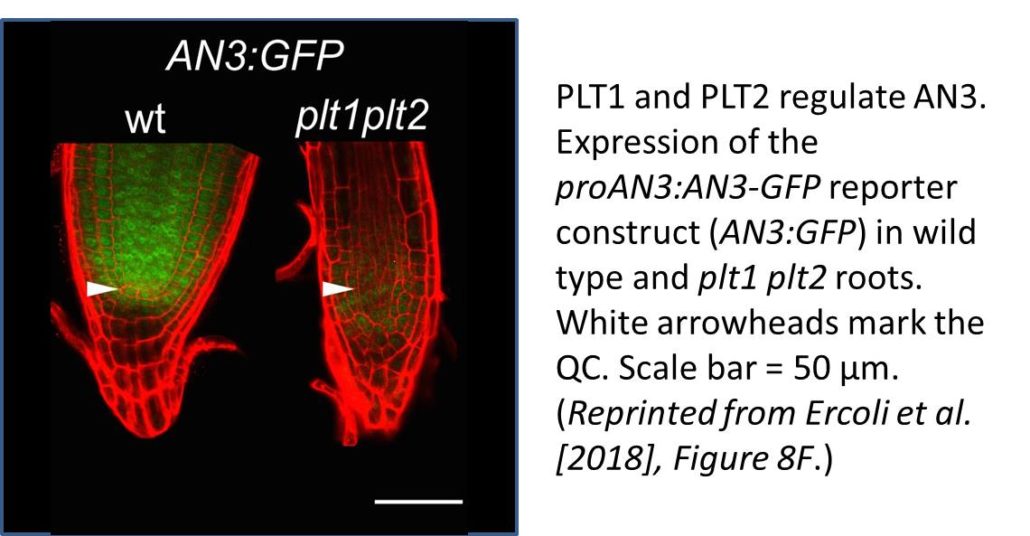
QC identity is specified by two parallel pathways directed by the transcription factors PLETHORA (PLT) and SCARECROW (SCR). Peak PLT expression determines the stem cell niche, while lower expression levels promote cell proliferation in the root meristem. Indeed, the expression patterns of PLT genes, especially PLT1, were altered in these lines: their expression zones were expanded in the root meristems of both GIF mutants and miR396 overexpressors but reduced in the QC only in GIF mutants. Unlike an3, the plt1 and an3 plt1 mutants had small root meristems, suggesting that the expanded expression of PLT1 in an3 is at least partially responsible for its enlarged root meristem. Chromatin immunoprecipitation (ChIP) assays showed that AN3 directly regulates PLT1. The PLTs regulate AN3 as well, as AN3 was strongly downregulated in double mutants derived from a cross between an AN3 reporter line and plt1 plt2 (see figure). AN3 also regulates SCR, as shown by analysis of reporter lines and previous ChIP-seq results. Therefore, in addition to controlling meristem length and QC organization, the GIFs help maintain the precise expression patterns of key developmental regulators in the root meristem, making them design stars, at least in the plant world.
REFERENCES
Ercoli, M.F., Ferela, A., Debernardi, J.M., Perrone, A.P., Rodriguez, R.E., and Palatnik. J.F. (2018). GIF transcriptional co-regulators control root meristem homeostasis. Plant Cell 30: http://www.plantcell.org/content/early/2018/01/18/tpc.17.00856
Heyman, J., Kumpf, R.P., and De Veylder, L. (2014). A quiescent path to plant longevity. Trends Cell Biol. 24: 443-448.
Horiguchi, G., Kim, G.T., and Tsukaya, H. (2005). The transcription factor AtGRF5 and the transcription coactivator AN3 regulate cell proliferation in leaf primordia of Arabidopsis thaliana. Plant J 43: 68-78.