Crossing the gap: Decrypting the genome facilitates gene identification in wild rice
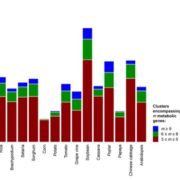
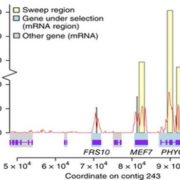
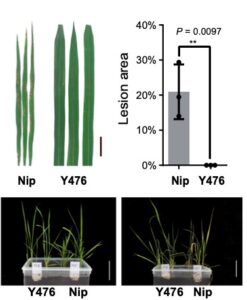 Domesticated crops provide a reliable food source but generally have little genetic diversity to cope with environmental fluctuations. By decrypting the genome of wild species such as the wild rice Oryza rufipogon, we may identify additional genetic variation useful for breeding process. However, the genomic heterozygosity of wild species is very challenging to work with. To cross this information gap, Huang et al. performed gapless assembly and comprehensive annotation of the wild rice Y476 genome. They incorporated segments of chromosomes from Y476 into the cultivated rice variety Nipponbare, allowing them to identify 254 Quantitative Trait Loci (QTLs) linked to agronomic traits and stress responses. Notably, a receptor-like kinase gene enhancing rice blast resistance was cloned from wild rice. This study provides a high-quality reference genome and establishes a platform for identifying beneficial genes in wild rice. (Summary by Yueh Cho @YuehCho1984) Nature Comms. 10.1038/s41467-024-48845-6.
Domesticated crops provide a reliable food source but generally have little genetic diversity to cope with environmental fluctuations. By decrypting the genome of wild species such as the wild rice Oryza rufipogon, we may identify additional genetic variation useful for breeding process. However, the genomic heterozygosity of wild species is very challenging to work with. To cross this information gap, Huang et al. performed gapless assembly and comprehensive annotation of the wild rice Y476 genome. They incorporated segments of chromosomes from Y476 into the cultivated rice variety Nipponbare, allowing them to identify 254 Quantitative Trait Loci (QTLs) linked to agronomic traits and stress responses. Notably, a receptor-like kinase gene enhancing rice blast resistance was cloned from wild rice. This study provides a high-quality reference genome and establishes a platform for identifying beneficial genes in wild rice. (Summary by Yueh Cho @YuehCho1984) Nature Comms. 10.1038/s41467-024-48845-6.