Gene Expression Study Reveals Distinct Virulence Modules of Corn Smut Fungus
Lanver et al. investigate fungal gene expression during corn infection by Ustilago maydis. The Plant Cell (2018) https://doi.org/10.1105/tpc.17.00764
By Daniel Lanver and Regine Kahmann
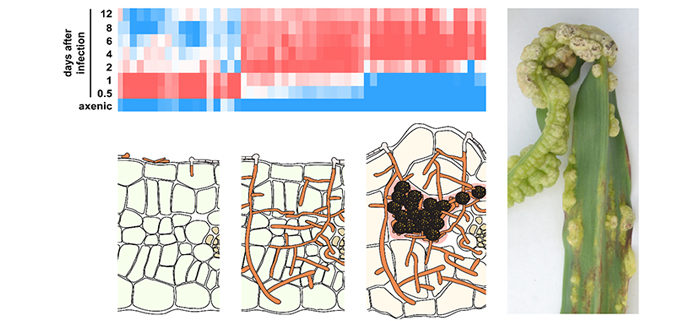
Background: The corn smut fungus Ustilago maydis is a model organism for studying how fungi can colonize living plant tissue. A special feature of this fungus is its ability to induce plant tumors, which become sites of massive fungal proliferation and spore formation. Work in the last decade has revealed that plant colonization by this fungus is largely governed by several hundred mostly novel secreted proteins. While the response of the maize plant to an infection by U. maydis has been studied in detail by looking at plant gene expression, a concomitant view of fungal gene expression was missing, largely because fungal biomass at early infection stages is very low.
Question: We wanted to determine the activity of all U. maydis genes during the entire fungal life cycle on and inside the plant to learn how the fungus deals with plant defense responses, which nutrients the fungus feeds on during colonization, and how plant tumors are induced. We tested this by deep mRNA sequencing.
Findings: Our comprehensive gene expression study has provided unexpected new leads concerning nutrition (hints that lipids and peptides are used), defense suppression and tumor induction during U. maydis colonization. The key finding of our study was that the activities of about one third of all U. maydis genes are co-regulated with the infection process. These genes can be divided into three distinct groups: one group of genes was active when the fungus was breaching the plant surface, one group of genes was particularly active early after plant colonization when the plant immune system is downregulated, and one group of genes was active only during the formation of tumors. This makes it very likely that the respective gene products contribute to the establishment of these discrete developmental stages. We also identified candidate regulators of these three so-called virulence modules and showed for one of the regulators of the tumor module that mutants lacking this regulator colonize plants but are unable to induce tumors in leaves.
Next steps: The identification of regulators for the specific modules will allow a system approach to the regulatory network controlling disease. Particularly attractive are the tumor module and its regulators, because they will enable the identification of genes responsible for inducing plant tumors, a prerequisite for elucidating how this is mechanistically achieved.
Daniel Lanver, André N. Müller, Petra Happel, Gabriel Schweizer, Fabian B. Haas, Marek Franitza, Clément Pellegrin, Stefanie Reissmann, Janine Altmüller, Stefan A Rensing, Regine Kahmann. (2018). The biotrophic development of Ustilago maydis studied by RNAseq analysis. Plant Cell. Published January 2018 https://doi.org/10.1105/tpc.17.00764.
Key words: virulence, fungi, plant tumors, gene expression