Convergent recruitment of TALE homeodomain life cycle regulators to direct sporophyte development in land plants and brown algae (eLIFE)
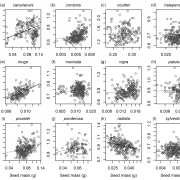
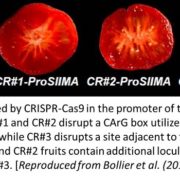
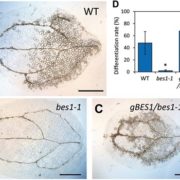
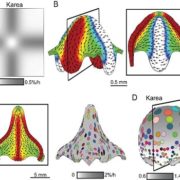
 Life cycles in sexually reproducing plants and algae alternate between diploid (sporophytic) and haploid (gametophytic) generations. The haploid gametophyte produces gametes that mate to generate the diploid sporophyte, which in turn undergoes meiosis to generate haploid spores. Development must be coordinated with these life cycle-dependent changes in ploidy, so that mating and meiosis take place in the correct generation. How is this coordination achieved? Arun et al. studied this question in Ectocarpus, a brown alga well suited for genetic analysis of life cycle transitions. Normally, gamete fusion (mating) marks the beginning of the sporophyte generation. However, in Ectocarpus unmated gametes may also switch to the sporophytic fate. The haploid origin of these so-called “partheno-sporophytes” facilitates analysis of loss-of-function mutations in genes that act in the diploid sporophyte. Using this system, they identified two genes, OUROBOROS and SAMSARA, without which the would-be partheno-sporophyte develops into a fully functional gametophyte. Both OUR and SAM encode three amino acid extension homeodomain (TALE HD) transcription factors needed to turn on the sporophyte transcriptional program. Interestingly, the same TALE HD is found in transcription factors that activate the zygotic program in Chlamydomonas and sporophyte development in Physcomitrella, two green lineage organisms that are not closely related to brown algae. These finding point to a common, and ancient origin of the machinery controlling eukaryotic life cycle transitions. (Summary by Frej Tulin) 10.7554/eLife.43101
Life cycles in sexually reproducing plants and algae alternate between diploid (sporophytic) and haploid (gametophytic) generations. The haploid gametophyte produces gametes that mate to generate the diploid sporophyte, which in turn undergoes meiosis to generate haploid spores. Development must be coordinated with these life cycle-dependent changes in ploidy, so that mating and meiosis take place in the correct generation. How is this coordination achieved? Arun et al. studied this question in Ectocarpus, a brown alga well suited for genetic analysis of life cycle transitions. Normally, gamete fusion (mating) marks the beginning of the sporophyte generation. However, in Ectocarpus unmated gametes may also switch to the sporophytic fate. The haploid origin of these so-called “partheno-sporophytes” facilitates analysis of loss-of-function mutations in genes that act in the diploid sporophyte. Using this system, they identified two genes, OUROBOROS and SAMSARA, without which the would-be partheno-sporophyte develops into a fully functional gametophyte. Both OUR and SAM encode three amino acid extension homeodomain (TALE HD) transcription factors needed to turn on the sporophyte transcriptional program. Interestingly, the same TALE HD is found in transcription factors that activate the zygotic program in Chlamydomonas and sporophyte development in Physcomitrella, two green lineage organisms that are not closely related to brown algae. These finding point to a common, and ancient origin of the machinery controlling eukaryotic life cycle transitions. (Summary by Frej Tulin) 10.7554/eLife.43101
[altmetric doi=”10.7554/eLife.43101″ details=”right” float=”right”]