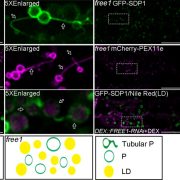
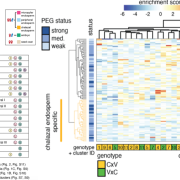
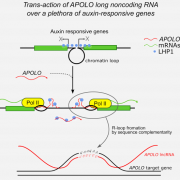
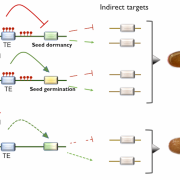
Combinations of maternal-specific repressive epigenetic marks in the endosperm control seed dormancy (bioRxiv.)
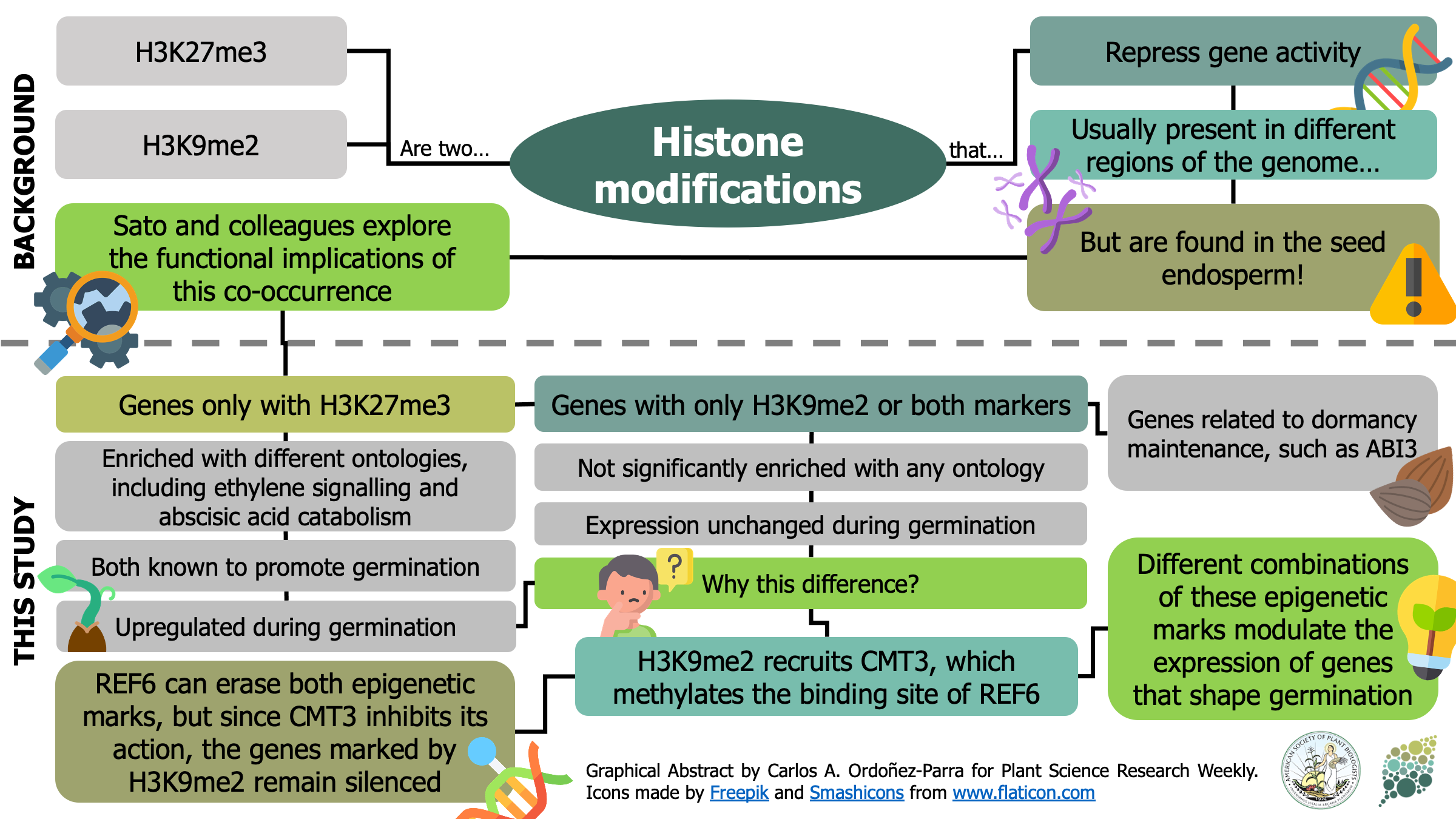
H3K27me3 [K27 (Lysine 27) trimethylation on histone H3] and H3K9me2 are two epigenetic modifications that repress gene activity in plants. While they are typically present in different genomic regions, both marks are found in the seed endosperm. Here, Sato and colleagues show that this histone modification duo is key to modulate gene activity during germination. Genes only marked with H3K27me3 were significantly enriched with gene ontologies including ethylene signaling and abscisic acid catabolism, both known for promoting germination. Moreover, they were found to be upregulated during germination process itself. Contrastingly, genes marked only with H3K9me2 or with both marks were not correlated to any ontology, nor had changes in their expression during germination. H3K9me2 recruits CHROMOMETHYL TRANSFERASE 3 (CMT3), which methylates the biding site of RELATIVE OF EARLY FLOWERING 6 (REF6) and inhibits its activity. Since REF6 can erase both H3K27me3 and H3K9me2 marks, CMT3 keeps marked genes inactive. Among the genes that present these two epigenetic marks, the authors found different dormancy maintenance genes such as ABA INSENSITIVE 3. Therefore, this research provides fascinating insights about how different combinations of epigenetic marks in the endosperm orchestrates the activation or silencing of genes that shape the germination process. (Summary by Carlos A. Ordóñez-Parra @caordonezparra) bioRxiv. 10.1101/2020.11.10.376806