Closing Gaps and Opening New Avenues During Megasporogenesis of Rice
The formation of the mature female gametophyte is one of the most complex processes in plants. It requires successive rounds of diploid mitosis, followed by meiosis and finally haploid mitosis, not to mention cell differentiation and cell degeneration in the multicellular gametophyte. Since the female gametophyte number and fertility directly affect seed yield and plant fitness, the identification of the main molecular programs controlling these important traits is an active field of research.
Unlike the male gametophyte, the female gametophytic tissue is scarce and surrounded by sporophytic-ovule tissues. These limitations have slowed our progress in understanding the molecular programs organizing the mature female gametophyte. The first part of this complex process occurs in diploid tissues (megasporogenesis) through mitotic divisions leading initially to the archesporial cell (AC) and finally, to the single cell that undergoes meiosis, the megaspore mother cell (MMC) that originates four haploid megaspores. In the second part, called megagametogenesis, three of the four haploid cells degenerate and one differentiates into the functional megaspore (FM), which then undergoes haploid mitosis and differentiation to generate the multicellular female gametophyte (Grossniklaus and Schneitz, 1998).
Several labs have made notable efforts, mainly in dicot plants, to study this process (Schmidt et al., 2011; Hafidh et al., 2016). However, little is known in cereals and even less at early stages of rice (Oryza sativa L.) gamete development (Kubo et al., 2013; Wu et al., 2015).
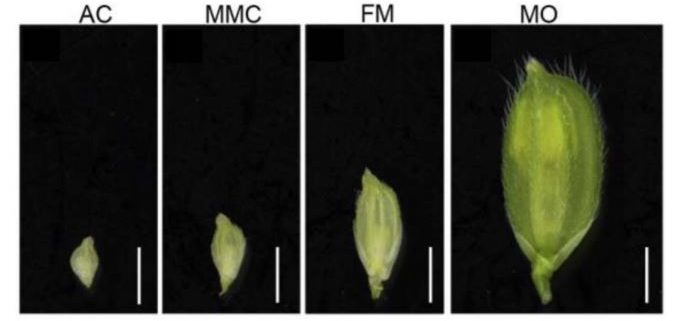
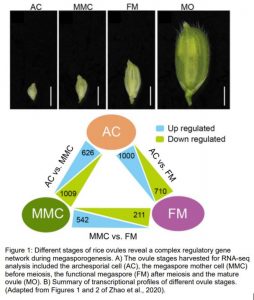 In this issue of Plant Physiology, Zhao et al. (2020) report a detailed transcriptomic experiment to understand the dynamics of the molecular programs controlling megasporogenesis in rice (Zhao et al., 2020). Since rice spikelet length correlates with the ovule developmental stage (Zeng et al., 2009), the authors evaluated the transcriptomic profiles of young ovules at three stages, the AC, the MMC before meiosis, and FM after meiosis. To collect the different ovule stages, the authors performed a meticulous ontological characterization of the developing flower followed by confocal microscopy of the female gametophyte structure.
In this issue of Plant Physiology, Zhao et al. (2020) report a detailed transcriptomic experiment to understand the dynamics of the molecular programs controlling megasporogenesis in rice (Zhao et al., 2020). Since rice spikelet length correlates with the ovule developmental stage (Zeng et al., 2009), the authors evaluated the transcriptomic profiles of young ovules at three stages, the AC, the MMC before meiosis, and FM after meiosis. To collect the different ovule stages, the authors performed a meticulous ontological characterization of the developing flower followed by confocal microscopy of the female gametophyte structure.
The transcriptomic analysis reported 1) a large number of differentially expressed genes during megasporogenesis; 5,274 up-regulated genes from a total of ≈ 23,000 rice genes; 2) preferential expression of genes at specific stages, i.e. up-regulation of 461 AC-preferential genes vs. 351 MMC-preferential genes and a smaller group of 146 up-regulated genes at both AC and MMC; 3) identification of transcription factor genes, some of them were not known to be involved in the process; and finally, 4) activation of specific molecular pathways related to auxin, plant–pathogen interaction and EPF-ERECTA-MAPK. The authors also provided further in silico and experimental data supporting a role for the auxin and receptor-like kinase pathways in the regulation of rice megasporogenesis.
To support the transcriptomic data, Zhao et al. validated the expression of nine genes using in situ hybridization. They confirmed specific expression for the nine genes at particular stages of rice ovule development, in agreement with the RNA-seq data. Within this group of selected genes, OsERECTA2 (OsER2) showed preferential expression at AC- and MMC-stages, whereas OsDEFL53, encoding a defensin-like peptide, was detected specifically at the AC stage. The in situ hybridization experiments support the idea that this dataset has a deep spatiotemporal resolution for expression analysis in rice with the potential to explore other responses taking place during these stages.
The authors also performed a functional analysis on OsER2, encoding an ERECTA-family receptor, using the CRISPR/Cas9 system. Previously, a mutant in this gene had been evaluated in a different rice cultivar. The mutant had stunted growth and reduced panicle size but the gametophytes were not analyzed (Zhang et al., 2018). Zhao et al. characterized their own oser2 mutant lines and found 1) that low grain yield accompanied the reduced plant height and panicle size; 2) normal pollen viability; and 3) aberrant female gametophyte structures. A further characterization of the female gametophyte of the oser2 mutant showed defects during germline specification with more than one AC-like or MMC-like cell at those early stages. Together, these results indicate that OsER2 affects seed set by controlling female germline cell specification.
This enormous dataset it is now publicly available at the NCBI BioProject repository, and, the authors encourage experimental validation of gene profiles since they cannot rule out that differentially expressed genes come from sporophytic surrounding tissues such as integuments and nucellus and not necessarily from megasporogenesis.
This experimental effort comes at a time when comparative transcriptomic and comparative physiology aiming to identify gene regulatory networks is blooming (Jones and Vandepoele, 2020). As an example, recent studies showed that the hormonal control of early ovule development in Arabidopsis thaliana and Solanum lycopersicum is differentially orchestrated by gibberellins and brassinosteroids (Barro-Trastoy et al., 2020). This has implications for comparative studies between model plants and crops/cereals but also for studies with an evolutionary perspective. The detailed map of gene profiles traced in this work goes beyond the identification of putative megasporogenesis candidate genes. It also provides information about other processes that are simultaneously occurring in the developing ovule, such as cell division and cell elongation, ovule nutrition, cell homeostasis, redox- and defense-related regulation, and cell wall softening. This study is therefore relevant for a broader group of plant biologists and has the potential to become both a powerful tool to understand the conservation of gene modules during early stages of the ovule development in plants and an effective tool for gene discovery.
Javier Edgardo Moreno
ORCID ID: 0000-0001-9763-5325
Instituto de Agrobiotecnología del Litoral (UNL-Conicet), Facultad de Bioquímica y Ciencias Biológicas, Universidad Nacional del Litoral
Santa Fe 3000, Argentina
Literature cited
Barro-Trastoy D, Carrera E, Banos J, Palau-Rodriguez J, Ruiz-Rivero O, Tornero P, Alonso JM, Lopez-Diaz I, Gomez MD, Perez-Amador MA (2020) Regulation of ovule initiation by gibberellins and brassinosteroids in tomato and Arabidopsis: two plant species, two molecular mechanisms. Plant J
Grossniklaus U, Schneitz K (1998) The molecular and genetic basis of ovule and megagametophyte development. Semin Cell Dev Biol 9: 227-238
Hafidh S, Fila J, Honys D (2016) Male gametophyte development and function in angiosperms: a general concept. Plant Reprod 29: 31-51
Jones DM, Vandepoele K (2020) Identification and evolution of gene regulatory networks: insights from comparative studies in plants. Curr Opin Plant Biol 54: 42-48
Kubo T, Fujita M, Takahashi H, Nakazono M, Tsutsumi N, Kurata N (2013) Transcriptome analysis of developing ovules in rice isolated by laser microdissection. Plant Cell Physiol 54: 750-765
Schmidt A, Wuest SE, Vijverberg K, Baroux C, Kleen D, Grossniklaus U (2011) Transcriptome analysis of the Arabidopsis megaspore mother cell uncovers the importance of RNA helicases for plant germline development. PLoS Biol 9: e1001155
Wu Y, Yang L, Cao A, Wang J (2015) Gene expression profiles in rice developing ovules provided evidence for the role of sporophytic tissue in female gametophyte development. PLoS One 10: e0141613-e0141613
Zeng YX, Hu CY, Lu YG, Li JQ, Liu XD (2009) Abnormalities occurring during female gametophyte development result in the diversity of abnormal embryo sacs and leads to abnormal fertilization in indica/japonica hybrids in rice. J Integr Plant Biol 51: 3-12
Zhang Y, Li S, Xue S, Yang S, Huang J, Wang L (2018) Phylogenetic and CRISPR/Cas9 studies in deciphering the evolutionary trajectory and phenotypic impacts of rice ERECTA genes. Front Plant Sci 9: 473