Callose deposition during pollen development
Madeleine Seale
University of Oxford
[email protected]
Callose is a cell wall component that is dynamically deposited and degraded during pollen development. Thanks to a new paper investigating pollen formation in cotton (Gossypium hirsutum), we now know that a pollen-specific protein regulates callose deposition by inhibiting the action of a transcription factor, WRKY15 (Li et al., 2020).
Callose is a polymer formed primarily of glucose units connected by β1-3-linkages. It is transiently deposited at sites of wounding, at plasmodesmata and pollen cell walls. Callose is also frequently present as a transient wall component in newly formed cell plates during cytokinesis of mitotically dividing plant cells (Scherp et al., 2001).
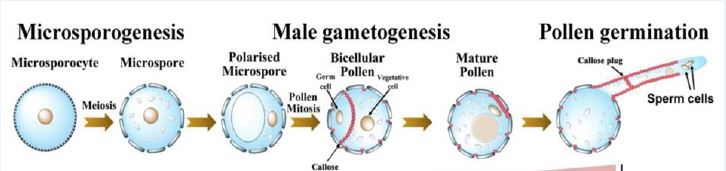
In the anthers of many angiosperms, a transient callose-rich cell wall is laid down, surrounding and separating each microsporocyte and callose is also deposited on the outer pollen wall (Blackmore et al., 2007; De Storme & Geelen, 2013). Callose is thought to isolate microspores from one another and is considered an impermeable barrier. Additionally, in many species that form these callose-rich walls, mutants with defective callose production do not create viable pollen (De Storme & Geelen, 2013). Nevertheless, the essential role of callose is somewhat contested as there is some evidence that large molecules can still cross the callose wall and some species do not deposit callose at this early meiotic stage (Scott et al., 2004). Despite this, callose deposition during meiosis has been observed across land plants including in mosses, liverworts and hornworts and lycophytes (Flowers 2018).
Li et al. (2020) have identified a gene expressed in cotton pollen, POLLEN SPECIFIC PROTEIN 231 (PSP231). They uncovered a sequence of regulatory interactions leading from PSP231 through to callose deposition phenotypes. PSP231 is part of the SKU5-SIMILAR (SKS) family of genes, which are frequently glycosylated but otherwise have unknown biochemical functions. Some SKS genes are involved in root development, and others have been shown to be expressed in pollen (Albani et al., 1992; Wittink et al., 2000; Zhou, 2019b).
Studying gametogenesis is notoriously difficult as mutants are frequently sterile and it is even more difficult in species such as Gossypium, in which established tools for pollen germination research are not yet available. Despite this, the authors persevered by using in vitro methods as well as RNAi and heterologous expression.
The authors found that psp231 RNAi lines were defective in male gametogenesis, forming pollen grains that were frequently withered and misshapen. Overexpressing PSP231 in Nicotiana tabacum pollen also resulted in reduced pollen germination. These findings illustrated an essential role for PSP231 and suggested that its expression had to be carefully balanced to achieve its correct effects.
To understand what PSP231 might be doing in cotton, the researchers analysed the transcriptome of wild type and PSP231-RNAi pollen. They found that two callose synthase genes were downregulated in the knockdown line compared to wild type. Furthermore, in the pollen of the tobacco PSP231 overexpressors, aniline blue staining highlighted increased callose deposition both in the newly formed wall dividing the vegetative and germ cells and in mature pollen.
In the absence of PSP231, GhWRKY15, showed increased expression. The GhWRKY15 protein is a transcriptional repressor and was able to bind to the promoters of the two callose synthase genes, consistent with the decreased expression levels of these genes.
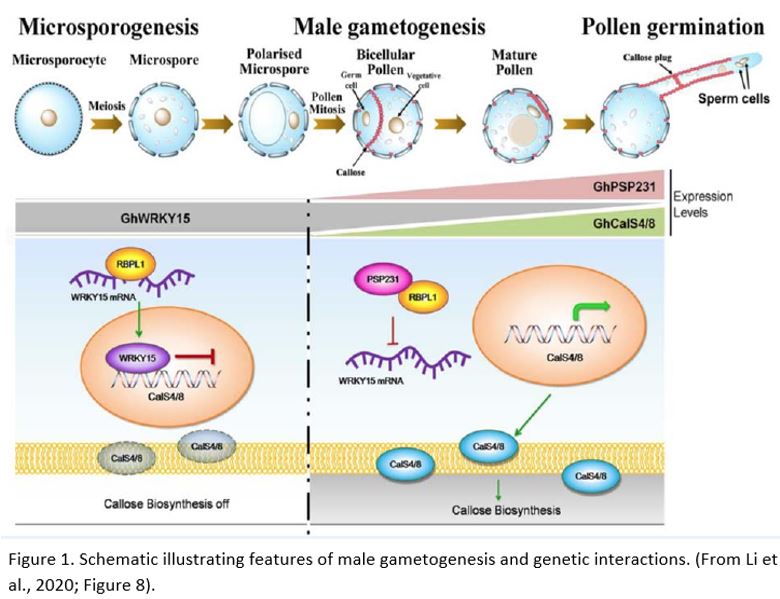 Li et al. (2020) also used a variety of in vitro and in vivo methods to establish that PSP231 forms a protein-protein complex with an RNA-binding protein, GhRBPL1. To complete the links in the chain, the authors then demonstrated that GhRBPL1 is able to specifically bind to the sense strand of GhWRKY15 mRNA. Taking the data together, they suggested a model in which PSP231 binds to and inhibits the actions of GhRBPL1. When de-repressed, GhRBPL1 is thought to bind to and stabilise GhWRKY15 mRNA. In turn, GhWRKY15 represses transcription of callose synthase genes. When PSP231 is present, GhWRKY15 is therefore downregulated and callose synthesis can take place (Fig. 1). As PSP231 is largely expressed at later stages of pollen development, it is likely that additional regulators also exist influencing the early-stage deposition of callose.
Li et al. (2020) also used a variety of in vitro and in vivo methods to establish that PSP231 forms a protein-protein complex with an RNA-binding protein, GhRBPL1. To complete the links in the chain, the authors then demonstrated that GhRBPL1 is able to specifically bind to the sense strand of GhWRKY15 mRNA. Taking the data together, they suggested a model in which PSP231 binds to and inhibits the actions of GhRBPL1. When de-repressed, GhRBPL1 is thought to bind to and stabilise GhWRKY15 mRNA. In turn, GhWRKY15 represses transcription of callose synthase genes. When PSP231 is present, GhWRKY15 is therefore downregulated and callose synthesis can take place (Fig. 1). As PSP231 is largely expressed at later stages of pollen development, it is likely that additional regulators also exist influencing the early-stage deposition of callose.
Remarkably, altered callose deposition was not only seen in plants, but also in Schizosaccharomyces pombe cells overexpressing PSP231. In fission yeast, dividing cells have callose in their new cell plates. When S. pombe overexpressed PSP231, cells were more often seen at late stages of cell division compared to wild-type, exhibiting callose-rich cell plates. These cells also showed growth retardation and decreased viability, likely due to the inability to degrade the excessively callosic septa.
While SKS-like genes have been known for some time, their roles have not been extensively studied. In Arabidopsis thaliana, SKU5 regulates root growth and mutants show twisted roots (Sedbrook et al., 2002). Similarly to PSP231, Arabidopsis sks mutants show modified cell geometry phenotypes with increased cell wall thickness in seedling roots (Zhou, 2019b). Previous research has demonstrated that SKS genes are frequently glycosylphosphatidylinositol (GPI)-anchored. Supporting this, Li et al. (2020) found that PSP231 is glycosylated. It is therefore likely that the PSP231 is anchored to lipid membranes within the cell as has been demonstrated for other SKS proteins (Sedbrook et al., 2002; Wittink et al., 2000; Zhou, 2019a). In Arabidopsis, disruption of GPI reduces the prevalence of GPI-anchored proteins including SKU5, and results in non-viable pollen and general disruption to cell wall synthesis (Stewart Gillmor et al., 2005). It will be interesting to see whether this type of post-translational modification is also essential for PSP231 function.
This is the one of the first instances in which a regulatory network with an SKS-like gene has been established and may open up further avenues of research for the roles of SKS genes in other species and tissues. The ability of PSP231 to substantially impact not only plant pollen development but also yeast cell division suggests that there may be common regulatory processes for callose synthesis across species and kingdoms. This has already been hinted at by the observation that both the plant and fungal genes that synthesise callose share significant sequence similarity (Latgé, 2007). The authors have established an important set of regulatory interactions governing the development and cellular structure of pollen, which will lead to a greater understanding of male reproductive development in plants.
Literature Cited
Albani, D., Sardana, R., Robert, L. S., Altosaar, I., Arnison, P. G., & Fabijanski, S. F. (1992). A Brassica napus gene family which shows sequence similarity to ascorbate oxidase is expressed in developing pollen. Molecular characterization and analysis of promoter activity in transgenic tobacco plants. The Plant Journal, 2(3), 331–342. https://doi.org/10.1046/j.1365-313X.1992.t01-32-00999.x
Blackmore, S., Wortley, A. H., Skvarla, J. J., & Rowley, J. R. (2007). Pollen wall development in flowering plants. New Phytologist, 174(3), 483–498. https://doi.org/10.1111/j.1469-8137.2007.02060.x
Chen, X.-Y., & Kim, J.-Y. (2009). Callose synthesis in higher plants. Plant Signaling & Behavior, 4(6), 489–492. https://doi.org/10.4161/psb.4.6.8359
De Storme, N., & Geelen, D. (2013). Cytokinesis in plant male meiosis. Plant Signaling & Behavior, 8(3), e23394. https://doi.org/10.4161/psb.23394
Latgé, J. P. (2007). The cell wall: A carbohydrate armour for the fungal cell. Molecular Microbiology, 66(2), 279–290. https://doi.org/10.1111/j.1365-2958.2007.05872.x
Scherp, P., Grotha, R., & Kutschera, U. (2001). Occurrence and phylogenetic significance of cytokinesis-related callose in green algae, bryophytes, ferns and seed plants. Plant Cell Reports, 20(2), 143–149. https://doi.org/10.1007/s002990000301
Scott, R. J., Spielman, M., & Dickinson, H. G. (2004). Stamen Structure and Function. The Plant Cell, 16(suppl 1), S46 LP-S60. https://doi.org/10.1105/tpc.017012
Sedbrook, J. C., Carroll, K. L., Hung, K. F., Masson, P. H., & Somerville, C. R. (2002). The Arabidopsis SKU5 Gene Encodes an Extracellular Glycosyl Phosphatidylinositol–Anchored Glycoprotein Involved in Directional Root Growth. The Plant Cell, 14(July), 1635–1648. https://doi.org/10.1105/tpc.002360.1996
Stewart Gillmor, C., Lukowitz, W., Brininstool, G., Sedbrook, J. C., Hamann, T., Poindexter, P., & Somerville, C. (2005). Glycosylphosphatidylinositol-anchored proteins are required for cell wall synthesis and morphogenesis in Arabidopsis. Plant Cell, 17(4), 1128–1140. https://doi.org/10.1105/tpc.105.031815
Wittink, F. R. A., Knuiman, B., Derksen, J., Čapková, V., Twell, D., Schrauwen, J. A. M., & Wullems, G. J. (2000). The pollen-specific gene Ntp303 encodes a 69-kDa glycoprotein associated with the vegetative membranes and the cell wall. Sexual Plant Reproduction, 12(5), 276–284. https://doi.org/10.1007/s004970050195
Zhou, K. (2019a). Glycosylphosphatidylinositol-Anchored Proteins in Arabidopsis and One of Their Common Roles in Signaling Transduction. Frontiers in Plant Science, 10(August), 1–20. https://doi.org/10.3389/fpls.2019.01022
Zhou, K. (2019b). GPI-anchored SKS proteins regulate root development through controlling cell polar expansion and cell wall synthesis. Biochemical and Biophysical Research Communications, 509(1), 119–124. https://doi.org/10.1016/j.bbrc.2018.12.081