BonnMu: a sequence-indexed resource of transposon-induced maize mutations for functional genomics studies (Plant Physiol.)
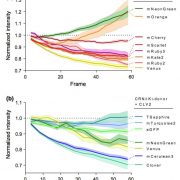
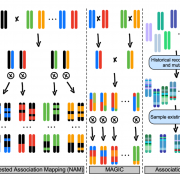
 Loss-of-function mutants have been invaluable tools in the geneticist’s toolbox for more than a century. More recently, libraries have been developed of knockout insertion mutants at known sites. Here, a new Mu-transposon insertion sequence-indexed maize resource is described. This new collection, BonnMu (developed in Bonn, Germany) complements the North American UniformMu and the Asian ChinaMu resources. A comparison of the BonnMu data from the B73 line and the UniformMu from the W22 line shows an abundance of conserved insertion hotspots between different genetic backgrounds. Furthermore, most insertions from both datasets were close to the transcription start site, which corresponds to open chromatin. The BonnMu collection provides easy-to-view images of segregating F2 populations that can be accessed from the genome browser, https://www.maizegdb.org/gbrowse. From the genome browser view, click “Select track” and under “Insertions” select “BonnMu insertions”. Go back to the browser and the insertion sites become visible in the locus. Clicking on the insertion site links to the Mu insertion identifiers (e.g., BonnMu0000812) and the respective F2–Mu-seq family (e.g., BonnMu-2-A-0596) with an image of its phenotype 10 days after germination. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00478
Loss-of-function mutants have been invaluable tools in the geneticist’s toolbox for more than a century. More recently, libraries have been developed of knockout insertion mutants at known sites. Here, a new Mu-transposon insertion sequence-indexed maize resource is described. This new collection, BonnMu (developed in Bonn, Germany) complements the North American UniformMu and the Asian ChinaMu resources. A comparison of the BonnMu data from the B73 line and the UniformMu from the W22 line shows an abundance of conserved insertion hotspots between different genetic backgrounds. Furthermore, most insertions from both datasets were close to the transcription start site, which corresponds to open chromatin. The BonnMu collection provides easy-to-view images of segregating F2 populations that can be accessed from the genome browser, https://www.maizegdb.org/gbrowse. From the genome browser view, click “Select track” and under “Insertions” select “BonnMu insertions”. Go back to the browser and the insertion sites become visible in the locus. Clicking on the insertion site links to the Mu insertion identifiers (e.g., BonnMu0000812) and the respective F2–Mu-seq family (e.g., BonnMu-2-A-0596) with an image of its phenotype 10 days after germination. (Summary by Mary Williams @PlantTeaching) Plant Physiol. 10.1104/pp.20.00478