Experimental Reproducibility 101 (Part 2)
This article is the second of three and is based on a workshop called “Reproducibility for all” presented at PlantBio18 by Benjamin Schwessinger, Sonali Roy, and Lenny Teytelman. In Part 1 (here), the authors describe the importance of having a data management plan, and the value of electronic notebooks.
3. Include a lot of detail when writing a protocol and share via the right platform.
Problems of reproducibility are often compounded by current scientific publishing policies. In many cases the information required to perform an experiment is insufficiently described in the methods section of a manuscript due to a set word limit, so essential details are missing. Or, when you try to find more information you hit a paywall and the information is held ransom to a subscription!
How can we overcome this problem? To begin with, by not contributing to it at all!
As this author suggests, think of a protocol as a ‘self-contained’ scientific publication intended for a beginner to read and understand the context of the experiment. Write the steps chronologically, include as much detail as possible (including but not limited to duration/time per step, reagent amount, vendor name, catalogue number, expected result, software packages, safety information) and also have notes for tips and tricks that make the protocol work for you.
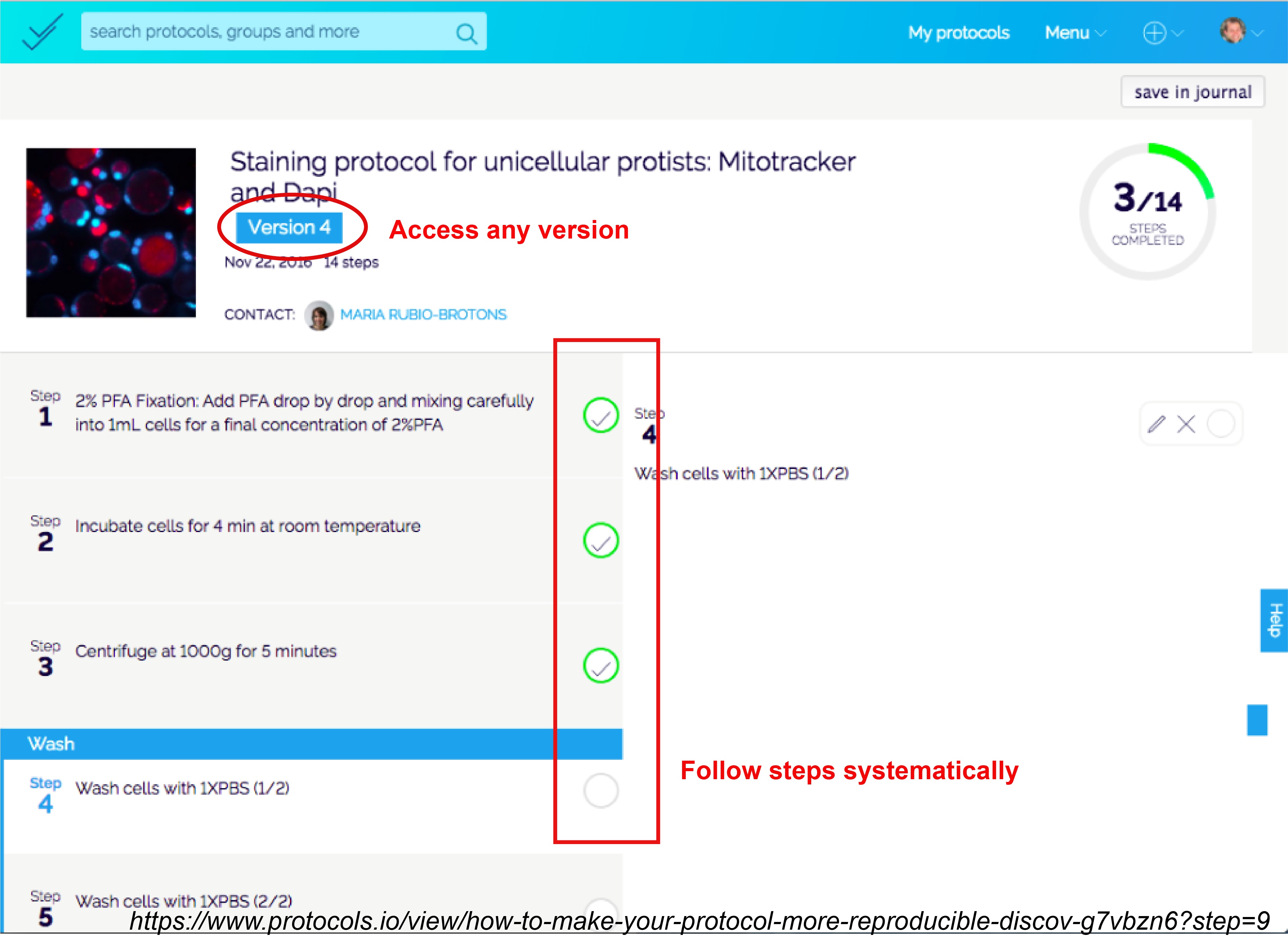
Hundreds of journals including eLife and PLOS now accept links to detailed methods on external websites such as Bioprotocols, protocols.io and JoVE (Journal of Visualized Experiments) that exclusively host protocols. Posting protocols not only bypasses the word limit a journal might have, but also these protocols can be accessed for free by individual users. Protocols are ordered and categorized with timed steps that are compatible with mobile apps, and different versions are available separately if changes are made to any one version.
4. Reagent repositories can help verify, curate, and distribute material you generate.
Situation three:
The signed Material Transfer Agreements (MTAs) have finally been mailed and you receive genetically modified bacterial strains from a highly regarded laboratory in your field! You immediately proceed to perform the large scale experiment you had planned over the next three months. Your more experienced colleague suggests you first sequence the strains to verify them.
The results were recently published in a reputable journal with an impact factor over 10 – do you really need to?
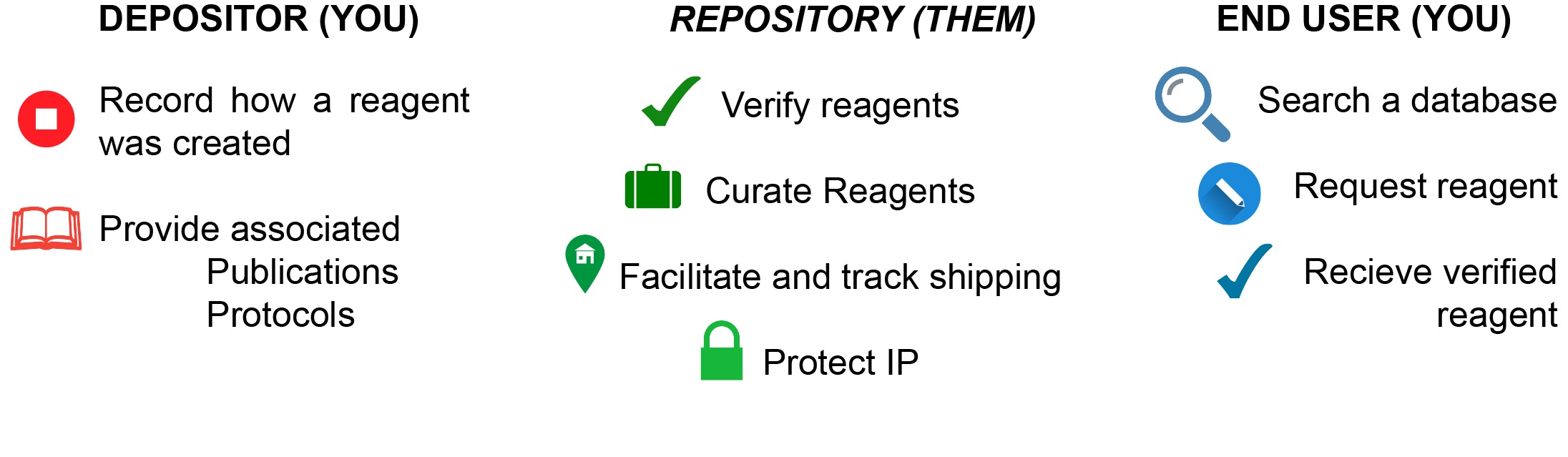
With a constant turnover of scientific personnel, most laboratories are often unable to keep track of all reagents and materials generated over time. Individual labs may lack the resources to properly organize, constantly validate or legally distribute these material. This adds to problems of reproducibility as researchers are forced to spend time recreating reagents or using misidentified tools.
Reagent repositories such as Addgene (plasmids), Arabidopsis Biological Resource Center (Arabidopsis seeds and stocks), American Type Culture Collection (microorganisms and cell lines) provide a practical solution. Scientists send material they generate to these repositories which verify the resources once received and archive the material for timely distribution upon request by users worldwide.
5. Deposit your data and code to public repositories ensuring access for future researchers.
Like ‘wet lab’ reagent repositories, data and code sharing repositories also provide organized, long-term access to your research. These repositories let users manage access to their files, licenses and provide persistent, unique and citable identifiers for their data such as Digital Object Identifiers (DOIs). Depositing valuable data or code to public repositories as opposed to personal websites could potentially help avoid an (unfortunately) common occurrence such as this:
You have found an exciting candidate gene in your newly sequenced model organism. You dig through published literature for data already available on it that would support your hypothesis about its function. This could possibly help you write your next grant! Luckily, you do find an article describing archived raw data about your gene and click the provided link.
“HTTP 404 Page not found. Please check the URL.”
Why share your data? Making data or code that is necessary to validate and/or reproduce your findings accessible upon publication is now mandatory. However, depositing valuable data onto public repositories also makes sure it will be available to future researchers, even when funding for a particular project/website/web-host runs out. Moreover, the more openly accessible your data/code is to researchers, the more widely it is used. This improves the citation rate for your work and consequently improves its ‘impact’ on that field.
What data should you share? Share data or code that cannot be easily generated or is valuable not only to fellow scientists but also to policy makers.
How should you share this data? Data repositories can be specific (funder/institution/discipline based) or general purpose. DataDryad, Figshare and Zenodo are examples of general purpose repositories. When sharing data, consider using permissive open source licenses such as the Creative Commons licence (data) or the MIT or Apache licence (code). These serve as legal agreements that allow users to freely modify and use a code as long as it acknowledged that the original author still holds the copyrights. See below for more information about some of these repositories.