APOPLASTP: prediction of effectors and plant proteins in the apoplast using machine learning ($)
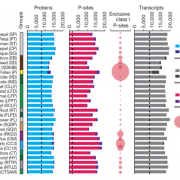
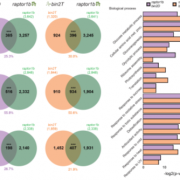
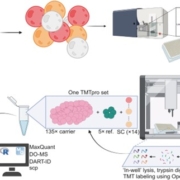
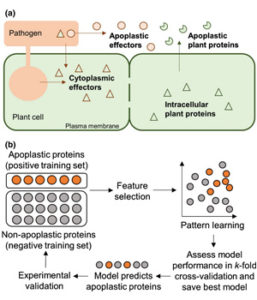 The apoplast represents a highly interactive site for intercellular communication and transport in plant-microbe interaction, that determines whether a pathogen can successfully overcome the early stages of plant defense. This site is also a battleground for an important class of apoplastic effectors that are known to perform exciting functions. Current methods of predicted apoplastic proteins are limited to microscopy and apoplastic proteomics which pose technical challenges. However, computational methods to predict the apoplastic proteins don’t exist. Sperschneider et al. have presented a machine learning platform called ApoplastP that predicts whether an effector or plant protein localizes to the apoplast. ApoplastP is based on common features of depletion in glutamic acid, and acidic/ charged amino acids. It has been validated by using numerous fungal effectors from the database and shows an accuracy of 75% with a false positive rate of 5%. Hence ApoplastP has started a pioneer computational method for predicting proteins with apoplastic localization and hence will promote functional studies on effectors and plant proteins in apoplastic space thereby understanding their role more rapidly. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14946
The apoplast represents a highly interactive site for intercellular communication and transport in plant-microbe interaction, that determines whether a pathogen can successfully overcome the early stages of plant defense. This site is also a battleground for an important class of apoplastic effectors that are known to perform exciting functions. Current methods of predicted apoplastic proteins are limited to microscopy and apoplastic proteomics which pose technical challenges. However, computational methods to predict the apoplastic proteins don’t exist. Sperschneider et al. have presented a machine learning platform called ApoplastP that predicts whether an effector or plant protein localizes to the apoplast. ApoplastP is based on common features of depletion in glutamic acid, and acidic/ charged amino acids. It has been validated by using numerous fungal effectors from the database and shows an accuracy of 75% with a false positive rate of 5%. Hence ApoplastP has started a pioneer computational method for predicting proteins with apoplastic localization and hence will promote functional studies on effectors and plant proteins in apoplastic space thereby understanding their role more rapidly. (Summary by Amey Redkar) New Phytol. 10.1111/nph.14946