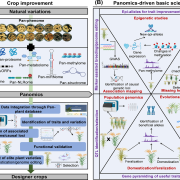
Updates on Resources, Software Tools, and Databases for Plant Proteomics in 2016–2017 ($) (Electrophoresis)
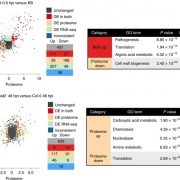
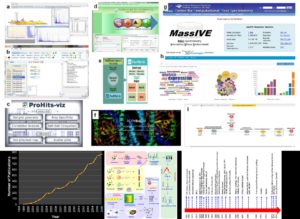 Proteomics, like metabolomics research, depends on the mass-spectrometry tools. Data processing, protein annotation, statistical and data analysis, as well as visualization are often the hurdles in obtaining meaningful biological insights from high throughput datasets. To this end, the author has summarized all open source community resources, software tools, and databases that surfaced in 2016-2017 and are useful for the plant proteomics community. Given that > 900 papers were published in plant proteomics in 2016 alone, better analysis tools are needed more than ever. In addition to compiling > 50 software tools for the plant proteomics community, the review describes publication trends in proteomics, a time-line showing important events in plant proteomics research, and a generic workflow for plant proteomics from sample to data. The review also includes a table categorizing the tools used for data preprocessing and analysis, statistical analysis, peptide identification, databases and spectral libraries, data visualization and interpretation. (Summary by Biswapriya Misra) Electrophoresis: 10.1002/elps.201700401
Proteomics, like metabolomics research, depends on the mass-spectrometry tools. Data processing, protein annotation, statistical and data analysis, as well as visualization are often the hurdles in obtaining meaningful biological insights from high throughput datasets. To this end, the author has summarized all open source community resources, software tools, and databases that surfaced in 2016-2017 and are useful for the plant proteomics community. Given that > 900 papers were published in plant proteomics in 2016 alone, better analysis tools are needed more than ever. In addition to compiling > 50 software tools for the plant proteomics community, the review describes publication trends in proteomics, a time-line showing important events in plant proteomics research, and a generic workflow for plant proteomics from sample to data. The review also includes a table categorizing the tools used for data preprocessing and analysis, statistical analysis, peptide identification, databases and spectral libraries, data visualization and interpretation. (Summary by Biswapriya Misra) Electrophoresis: 10.1002/elps.201700401