Arabidopsis Framework Model version 2 predicts the effects of circadian clock misregulation
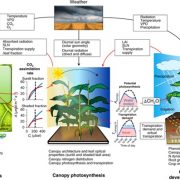
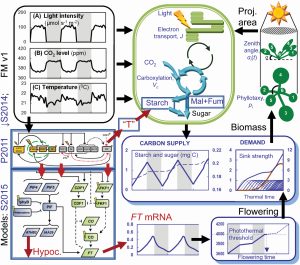 Experimental or “wet lab” biologists look at data, develop a hypothesis to explain it, test the hypothesis, and repeat. Through these approaches, our understanding of life is continually being expanded and refined. A complementary approach involves developing mathematical models to explain observations, and then testing those models by tweaking them and comparing the outputs to real-life experiments. In a new article, Chew et al. describe the results from a large-scale, multi-component computational model (“Framework Model 2”) that integrates models of photosynthesis and carbon supply, starch metabolism, growth, and the biological clock. The authors used “diverse metabolic and physiological data to combine and extend mathematical models of rhythmic gene expression, photoperiod-dependent flowering, elongation growth and starch metabolism”. Their model accurately predicted the phenotypic effects of several mutants affecting the circadian clock, demonstrating its usefulness in linking genotype to phenotype, and validating this approach to understand complex traits. (Summary by Mary Williams @PlantTeaching) in silico Plants 10.1093/insilicoplants/diac010
Experimental or “wet lab” biologists look at data, develop a hypothesis to explain it, test the hypothesis, and repeat. Through these approaches, our understanding of life is continually being expanded and refined. A complementary approach involves developing mathematical models to explain observations, and then testing those models by tweaking them and comparing the outputs to real-life experiments. In a new article, Chew et al. describe the results from a large-scale, multi-component computational model (“Framework Model 2”) that integrates models of photosynthesis and carbon supply, starch metabolism, growth, and the biological clock. The authors used “diverse metabolic and physiological data to combine and extend mathematical models of rhythmic gene expression, photoperiod-dependent flowering, elongation growth and starch metabolism”. Their model accurately predicted the phenotypic effects of several mutants affecting the circadian clock, demonstrating its usefulness in linking genotype to phenotype, and validating this approach to understand complex traits. (Summary by Mary Williams @PlantTeaching) in silico Plants 10.1093/insilicoplants/diac010