Another gun Dismantled: ABSCISIC ACID INSENSITIVE4 Is Not a Target of Retrograde Signaling
By
The nuclear genome encodes the majority of chloroplast-localized proteins and, in return, chloroplasts exert some control over nuclear gene expression via so-called retrograde signals. These signals derive from developing chloroplasts (referred to as biogenic signals) or from mature chloroplasts in response to environmental constraints such as excessive light, drought, or heat (referred to as operational signals; Crawford et al., 2018; Dubreuil et al., 2018; Hernández-Verdeja and Strand, 2018). The major nuclear targets of plastid signals are photosynthesis-associated nuclear genes (PhANGs); retrograde signals also target developmental processes such as seedling photomorphogenesis (Martín et al., 2016).
Despite the plethora of research focusing on chloroplast signaling, retrograde signals remain poorly understood and probably consist of several signals including reactive oxygen species and metabolites (Fig. 1). In most studies, biogenic retrograde signaling is induced by the inhibition of carotenoid biosynthesis using norflurazon or by the inhibition of plastid translation using lincomycin, which inhibit PhANG expression. Six Arabidopsis (Arabidopsis thaliana) genomes uncoupled (gun) mutants have been isolated that maintain PhANG expression under these treatments (Susek et al., 1993). All six original GUN proteins are localized in the chloroplast; GUN2 to GUN6 act in the tetrapyrrole synthesis pathway, and GUN1 is a pentatricopeptide repeat protein, responding to disruptions in plastid gene expression.
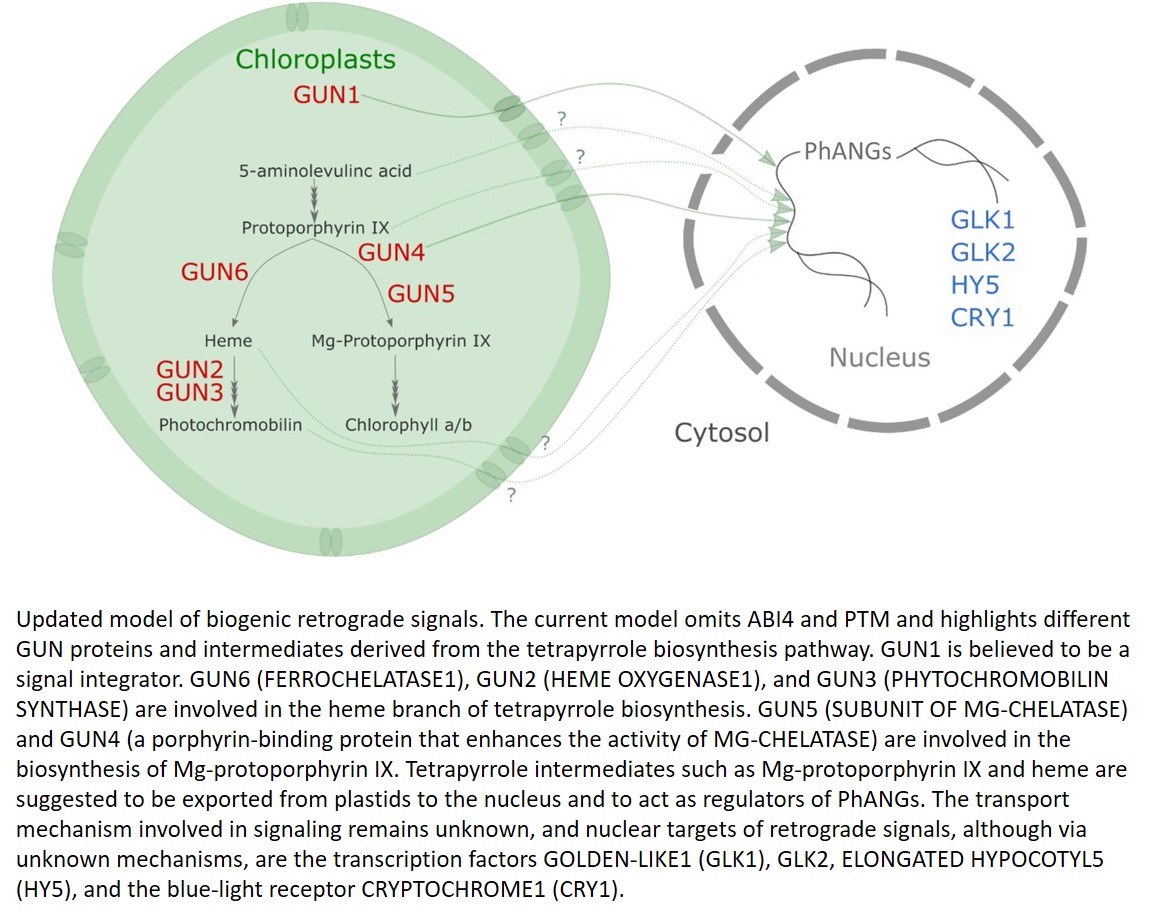
Updated model of biogenic retrograde signals. The current model omits ABI4 and PTM and highlights different GUN proteins and intermediates derived from the tetrapyrrole biosynthesis pathway. GUN1 is believed to be a signal integrator. GUN6 (FERROCHELATASE1), GUN2 (HEME OXYGENASE1), and GUN3 (PHYTOCHROMOBILIN SYNTHASE) are involved in the heme branch of tetrapyrrole biosynthesis. GUN5 (SUBUNIT OF MG-CHELATASE) and GUN4 (a porphyrin-binding protein that enhances the activity of MG-CHELATASE) are involved in the biosynthesis of Mg-protoporphyrin IX. Tetrapyrrole intermediates such as Mg-protoporphyrin IX and heme are suggested to be exported from plastids to the nucleus and to act as regulators of PhANGs. The transport mechanism involved in signaling remains unknown, and nuclear targets of retrograde signals, although via unknown mechanisms, are the transcription factors GOLDEN-LIKE1 (GLK1), GLK2, ELONGATED HYPOCOTYL5 (HY5), and the blue-light receptor CRYPTOCHROME1 (CRY1).
How the retrograde signaling is perceived by the nucleus is still enigmatic, and in this context, the transcription factor ABSCISIC ACID INSENSITIVE4 (ABI4) was proposed to act as a nuclear target of the GUN1-mediated retrograde signals (Nott et al., 2006; Koussevitzky et al., 2007; Sun et al., 2011). Several reports showed that abi4 mutants have a weak gun phenotype exhibiting elevated PhANG expression following norflurazon treatment. In addition, many photosynthesis-related nuclear genes controlled by plastid signals contain abscisic acid-responsive elements in their promoters (Koussevitzky et al., 2007). Surprisingly, several research groups were unable to reproduce the gun phenotype described for abi4 mutants. In this issue of Plant Physiology, Kacprzak et al. (2019) address this by analyzing the gun phenotypes in multiple abi4 alleles in different laboratories. The authors show that abi4 mutants, unlike gun1, failed to restore the expression of a set of PhANGs in the presence of norflurazon and lincomycin. Thus, Kacprzak et al. (2019) convincingly demonstrate that ABI4 is not a target of retrograde signaling and should be omitted from future models of biogenic retrograde signaling.
Interestingly, the same researchers last year dismantled another gun-like mutant. The plastid transmembrane-localized transcription factor PTM was once described as being activated by GUN1. After this activation, the cleaved PTM moved to the nucleus, where it promoted ABI4 expression and repressed PhANG expression. However, with an approach similar to that presented here, Page et al. (2017) concluded that PTM should be excluded as a major player in retrograde signaling. Based on these results, it would be interesting to further investigate the factors that might explain discrepancies with the original studies, especially in the case of abi4, since several groups previously reported a gun-like phenotype for abi4. Together, these studies finally resolved uncertainties surrounding the roles of ABI4 and PTM in retrograde signaling. They point to the need to have several lines of evidence supporting our conclusions and to carefully question the biological relevance of data.
The work of Kacprzak and colleagues proves that much work remains to be done to understand the complex organellar signaling events during development and stress responses. An important starting point would be to unravel the function of GUN1, the main mediator of retrograde signaling. Recent work has shown that GUN1 is involved in chloroplast biogenesis as well as retrograde signaling. Rapid protein turnover in green tissues helps to specify GUN1 accumulation during plastid development and stress (Song et al., 2018; Wu et al., 2018; Zhao et al., 2018). Nevertheless, it remains unknown how GUN1 degradation is prevented in the absence of functioning chloroplasts and what type of signal this releases. Future endeavors will help bridge our understanding of chloroplast damage, GUN1 accumulation, nuclear gene expression, and ultimately plant performance. To achieve this, we need a multidisciplinary approach and possibly many more guns to be dismantled.
REFERENCES
Crawford T, Lehotai N, Strand Å (2018) The role of retrograde signals during plant stress responses. J Exp Bot 69: 2783–2795
Dubreuil C, Jin X, Barajas-López JD, Hewitt TH, Tanz S, Dobrenel T, Schröder W, Hanson J, Pesquet E, Grönlund A, et al. (2018) Establishment of photosynthesis through chloroplast development is controlled by two distinct regulatory phases. Plant Physiol 176: 1199–1214
Hernández-Verdeja T, Strand Å (2018) Retrograde signals navigate the path to chloroplast development. Plant Physiol 176: 967–976
Kacprzak SM, Mochizuki N, Naranjo B, Xu D, Leister D, Kleine T, Okamoto H, Terry MJ (2019) Plastid-to-nucleus retrograde signalling during chloroplast biogenesis does not require ABI4. Plant Physiol.
Koussevitzky S, Nott A, Mockler TC, Hong F, Sachetto-Martins G, Surpin M, Lim J, Mittler R, Chory J (2007) Signals from chloroplasts converge to regulate nuclear gene expression. Science 316: 715–719
Martín G, Leivar P, Ludevid D, Tepperman JM, Quail PH, Monte E (2016) Phytochrome and retrograde signalling pathways converge to antagonistically regulate a light-induced transcriptional network. Nat Commun 7: 11431
Nott A, Jung HS, Koussevitzky S, Chory J (2006) Plastid-to-nucleus retrograde signaling. Annu Rev Plant Biol 57: 739–759
Page MT, Kacprzak SM, Mochizuki N, Okamoto H, Smith AG, Terry MJ (2017) Seedlings lacking the PTM protein do not show a genomes uncoupled (gun) mutant phenotype. Plant Physiol 174: 21–26
Song L, Chen Z, Larkin RM (2018) The genomes uncoupled mutants are more sensitive to norflurazon than wild type. Plant Physiol 178: 965–971
Sun X, Feng P, Xu X, Guo H, Ma J, Chi W, Lin R, Lu C, Zhang L (2011) A chloroplast envelope-bound PHD transcription factor mediates chloroplast signals to the nucleus. Nat Commun 2: 477
Susek RE, Ausubel FM, Chory J (1993) Signal transduction mutants of Arabidopsis uncouple nuclear CAB and RBCS gene expression from chloroplast development. Cell 74: 787–799
Wu GZ, Chalvin C, Hoelscher M, Meyer EH, Wu XN, Bock R (2018) Control of retrograde signaling by rapid turnover of GENOMES UNCOUPLED1. Plant Physiol 176: 2472–2495
Zhao X, Huang J, Chory J (2018) genome uncoupled1 mutants are hypersensitive to norflurazon and lincomycin. Plant Physiol 178: 960–964



