An Improved Tool for Mapping the Membrane-Associated Protein Interactome
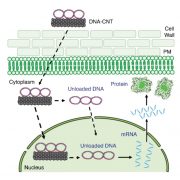
Protein-protein interactions mediate fundamental biological processes. Yeast two-hybrid assays can be used to detect protein-protein interactions in a fast and large-scale manner. However, in traditional yeast-two hybrid assays, the prey and bait proteins must be located in the nucleus to activate reporter genes. This requirement limits the application of this assay in detecting membrane protein interactions to using soluble domains that are small enough to enter the nucleus. Instead of using DNA-binding and activation domains of the traditional yeast two-hybrid system, split-ubiquitin yeast systems allow the detection of interactions between membrane proteins (Johnsson and Varshavsky, 1994; Stagljar et al., 1998). The earlier versions of split-ubiquitin systems allowed the fusion of membrane proteins directly to the N- and C-terminal parts of ubiquitin (split ubiquitin, Nub and Cub; Fig. 1A). Cub is usually fused with a transactivator complex PLV (for protein A-LexA-VP16; Fig. 1A). The interaction between membrane proteins brings together Nub and Cub, and this allows the cleavage and release of PLV by ubiquitin-specific proteases (USPs; Fig. 1B). Cleaved PLV is then translocated to the nucleus, where LexA serves as a DNA-binding domain and VP16 serves as a transcription activation domain to activate reporter genes (Fig. 1B). The mating-based split-ubiquitin system is further optimized to allow large-scale screening for membrane protein interactions with more stringent selection and easy gateway cloning (Obrdlik et al., 2004).
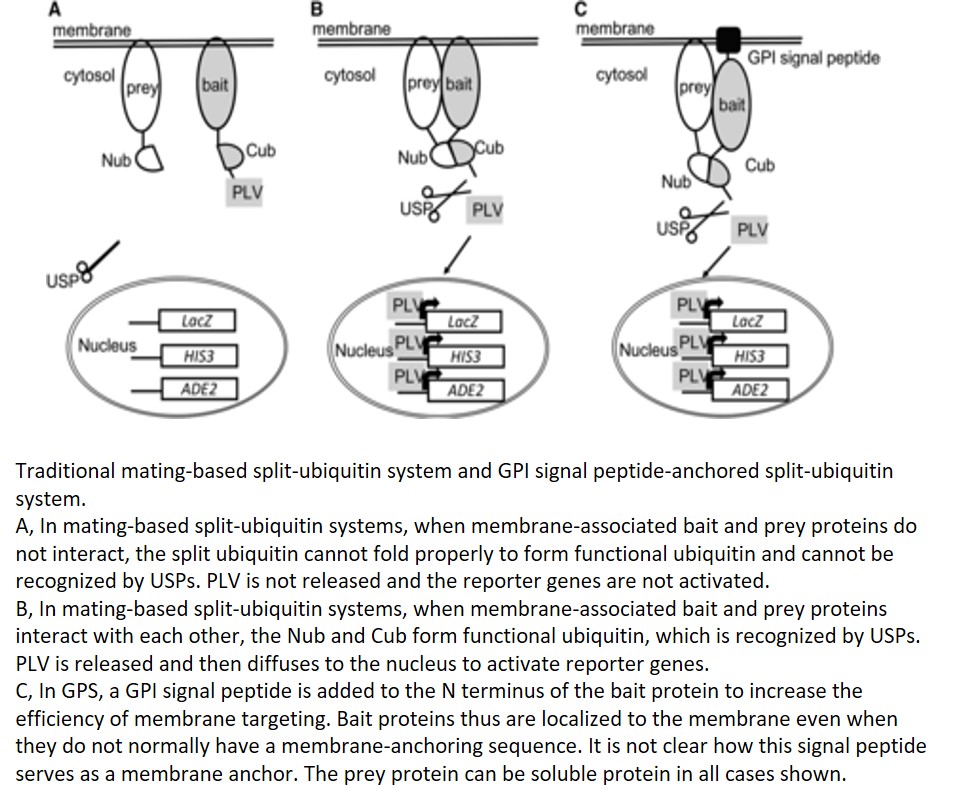
The prey in mating-based split-ubiquitin systems can be either soluble or membrane proteins fused with Nub, but the bait needs to be membrane associated to avoid autoactivation by diffusion of the fusion protein of Cub-PLV to the nucleus. Furthermore, mating-based split-ubiquitin systems require bait proteins to be anchored to the membrane using their N termini to ensure proper cleavage of PLV. Thus, membrane proteins that use the C terminus for membrane anchoring are generally not suitable as baits in this assay. In order to solve this problem, a membrane-anchoring protein, the 36-amino-acid OST4p subunit of the oligosaccharide transferase complex, has been used to target bait proteins to the membrane surface (Möckli et al., 2007). This strategy helps with membrane targeting of some soluble proteins but has limitations for truncated versions of the same proteins. In this issue of Plant Physiology, Zhang et al. make a further contribution to solving this problem by adding the GPI (for glycosyl-phosphatidylinositol) signal peptide from glucan-1,3-β-glucosidase 2 (Exg2) to the N terminus of the bait protein (Zhang et al., 2018; Fig. 1C). This signal peptide allows fusion proteins to be inserted in the endoplasmic reticulum membrane and transported through the vesicle trafficking pathway. Although it is not clear how this signal peptide serves as a membrane anchor and whether the protein is attached or inserted to the membrane. The new GPI signal peptide-anchored split-ubiquitin system has higher efficiency in targeting the soluble fragment of Sec11, comprising a small fragment of the N-terminal 149 residues of this protein, by comparison with the yeast OST4p leader sequence previously used for anchoring. It will be interesting to test the efficiency of this new system in targeting other proteins to the membrane.
About 30% of human genes and a similar percentage of plant genes encode membrane proteins (Almén et al., 2009). Dissecting the interaction networks of membrane proteins is critical for understanding the mechanisms by which these proteins function. The improved mating-based split-ubiquitin system reported by Zhang et al. makes it possible to map the membrane-associated protein interactome in a fast and efficient way. This novel tool will contribute greatly to our understanding of the mechanisms by which membrane proteins function. The authors will kindly provide the maps and vectors needed to use this system if you submit a request through their Web site, www.psrg.org.uk.
.
REFERENCES
Almén MS, Nordström KJ, Fredriksson R, Schiöth HB (2009) Mapping the human membrane proteome: a majority of the human membrane proteins can be classified according to function and evolutionary origin. BMC Biol 7: 50
Johnsson N, Varshavsky A (1994) Split ubiquitin as a sensor of protein interactions in vivo. Proc Natl Acad Sci USA 91: 10340–10344
Möckli N, Deplazes A, Hassa PO, Zhang Z, Peter M, Hottiger MO, Stagljar I, Auerbach D (2007) Yeast split-ubiquitin-based cytosolic screening system to detect interactions between transcriptionally active proteins. Biotechniques 42: 725–730
Obrdlik P, El-Bakkoury M, Hamacher T, Cappellaro C, Vilarino C, Fleischer C, Ellerbrok H, Kamuzinzi R, Ledent V, Blaudez D, et al. (2004) K+ channel interactions detected by a genetic system optimized for systematic studies of membrane protein interactions. Proc Natl Acad Sci USA 101: 12242–12247
Stagljar I, Korostensky C, Johnsson N, te Heesen S (1998) A genetic system based on split-ubiquitin for the analysis of interactions between membrane proteins in vivo. Proc Natl Acad Sci USA 95: 5187–5192
Zhang B, Karnik R, Donald N, Blatt MR (2018) A GPI signal peptide-anchored split-ubiquitin (GPS) system for detecting soluble bait protein interactions at the membrane. Plant Physiol 178: 13–17